Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Dr. Abriel’s work is supported by a grant of the Swiss National Science Foundation (310030_184783) and the University of Bern.
Na V 1.5 has been shown to interact with a growing list of proteins ( Fig. 18.1 ; Table 18.1 ). Mutations in genes encoding Na V 1.5 and several interacting proteins have been found in patients with inherited arrhythmias, such as congenital long QT syndrome (LQTS) and Brugada syndrome (BrS). Proteins interacting with Na V 1.5 have been classified as (1) anchoring or adaptor proteins involved in trafficking, targeting, and anchoring the channel to specific membrane compartments; (2) enzymes, such as protein kinases or ubiquitin ligases, interacting with and modifying the channel structure via posttranslational modifications ; and (3) proteins modulating the biophysical properties of Na V 1.5 (see Table 18.1 ). These classifications are not mutually exclusive. Na V 1.5 channels may also interact with each other (i.e., dimerize), likely mediated by the protein 14-3-3. ,
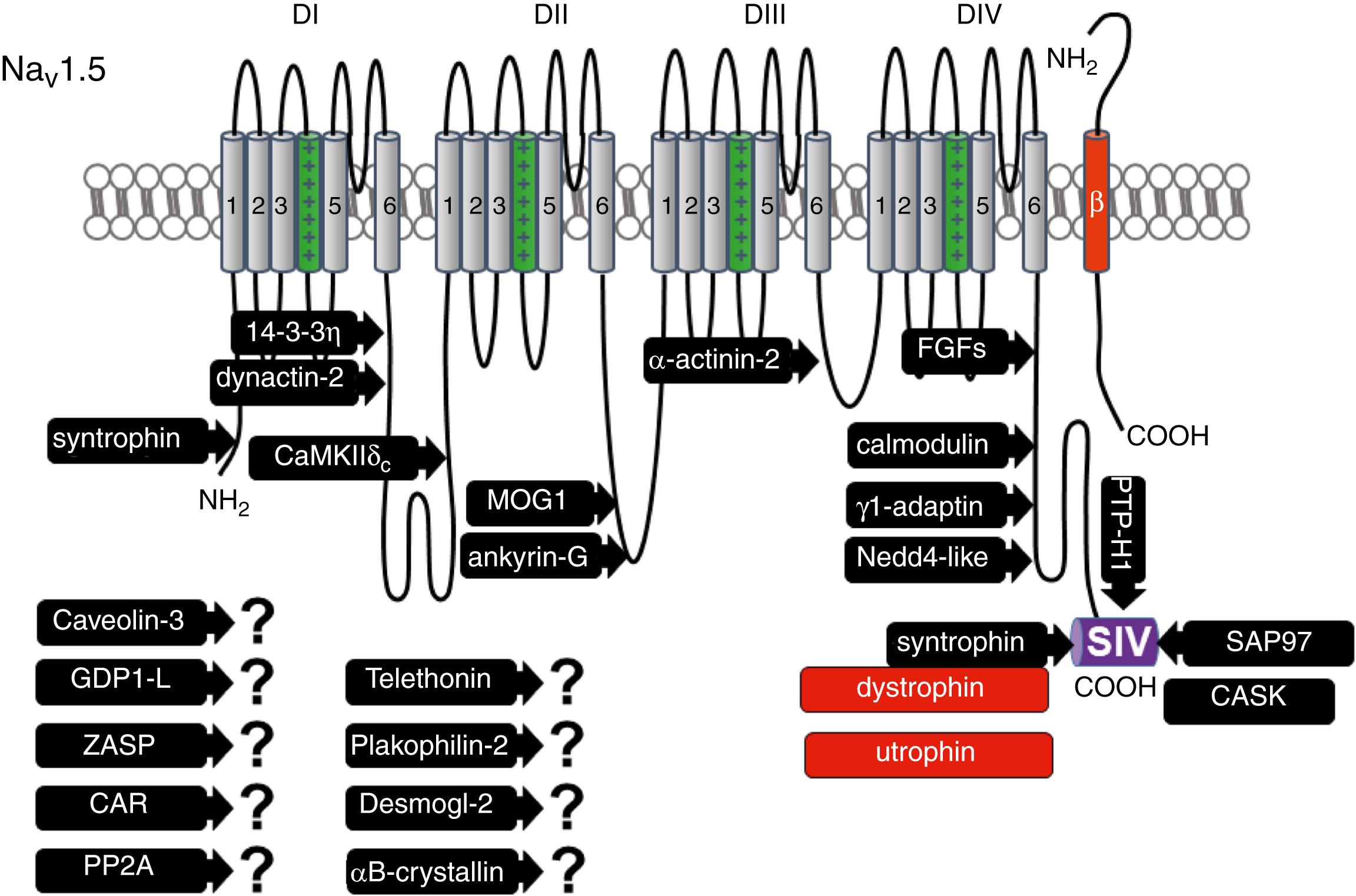
| Protein | Main Effects on Na V 1.5 | Mutated in Cardiac Disorder | Interaction Domain on Na V 1.5 | UniProt Reference | References |
|---|---|---|---|---|---|
| Anchoring Adaptor Proteins | |||||
| α-Actinin-2 | Involved in trafficking of Na V 1.5 by unknown mechanisms | Dilated and hypertrophic cardiomyopathy | Intracellular loop between DIII and IV | P35609 (ACTN2_HUMAN) | , , |
| αB-crystallin | Increase peak I Na , Na V 1.5 surface expression, reduction Na V 1.5 ubiquitination | Dilated cardiomyopathy and desmin-related myopathy | DII–III linker and C-terminus | P02511 (CRYAB_HUMAN) | , |
| γ1-Adaptin | Upregulation of I Na | N/A | Y1810 (C-terminus) | O43747 (AP1G1_HUMAN) | |
| Ankyrin-G | Na V 1.5 trafficking and anchoring to the cell membrane, in particular the intercalated discs | Na V 1.5 binding site mutated in one BrS patient | VPIA xx SD motif in intracellular loop DII–III | Q12955 (ANK3_HUMAN) | |
| CAR | Involved in Na V 1.5 trafficking by unknown mechanisms | N/A | Not determined | P78310 (CXAR_HUMAN) | |
| CASK | Downregulation of I Na | N/A | PDZ domain–binding motif in C-terminus | O14936 (CSKP_HUMAN) | |
| Desmoglein-2 | Overexpression of ACM mutants in mice reduces I Na | ACM | Not determined | Q14126 (DSG2_HUMAN) | |
| Dynactin-2 | Involved in Na V 1.5 trafficking by unknown mechanisms | N/A | Intracellular loop between DI and II | Q13561 (DCTN2_HUMAN) | |
| MOG1 | Involved in Na V 1.5 trafficking by unknown mechanisms | BrS | Intracellular loop between DII and III | Q9HD47 (MOG1_HUMAN) | , |
| SAP97 | Not yet determined | N/A | PDZ domain–binding motif in C-terminus | Q12959 (DLG1_HUMAN) | , |
| Syntrophin proteins | Adapt to dystrophin and utrophin complex and Na V 1.5 stabilization at myocyte lateral membrane | α1-Syntrophin gene ( SNTA1 ) mutated in LQTS and SIDS | PDZ domain–binding motif in C-terminus and internal PDZ-binding domain in N-terminus | Q13424 (SNTA1_HUMAN) | |
| Proteins That Affect INa Biophysical Properties | |||||
| 14-3-3η | Modulation of steady-state inactivation | N/A | PY-motif in intracellular loop between DI and II | Q04917 (1433F_HUMAN) | |
| Calmodulin | Many discrepant effects. May confer intracellular calcium sensitivity to Na V 1.5 May permit Na V 1.5 multimerization |
LQTS, ventricular tachycardia | IQ-motif (1900–1920) in C-terminus and DIII–IV linker (1510–1530 and 1490–1501) | P62158 (CALM_HUMAN) | , , , |
| Caveolin-3 | Mutant of caveolin-3 induces late current | LQTS, SIDS, and hypertrophic cardiomyopathy | Not determined | P56539 (CAV3_HUMAN) | , , , |
| FHF proteins | Modulation of steady-state inactivation, recovery from inactivation, and density at cell membrane | BrS | Residues 1773–1832 in C-terminus | P61328 (FGF12_HUMAN) | |
| Plakophilin-2 | Silencing reduces I Na , negatively shifts steady-state inactivation, and slows recovery from inactivation | ACM and BrS | Not determined | Q99959 (PKP2_HUMAN) | |
| Telethonin | Modulation of voltage dependence of activation | Hypertrophic cardiomyopathy | Not determined | O15273 (TELT_HUMAN) | , , |
| Enzymes | |||||
| CaMKIIδc | Phosphorylation of residues in intracellular loop I and modulation of biophysical properties | N/A | Intracellular loop between DI and II and via β IV -spectrin | Q13557 (KCC2D_HUMAN) | |
| GPD1-L | Loss-of-function variants reduce I Na by modulating PKC-dependent phosphorylation of Na V 1.5 | BrS | Not determined | Q8N335 (GPD1L_HUMAN) | |
| Nedd4-like E3 ubiquitin ligases | Ubiquitination and internalization | N/A | PY-motif in C-terminus | Q96PU5 (NED4L_HUMAN) | , |
| PP2A | Increase persistent I Na | N/A | Not determined | P67775 (PP2AA_HUMAN) | , |
| PTPH1 | Dephosphorylation (site unknown) and modulation of biophysical properties | N/A | PDZ-domain binding motif in C-terminus | P26045 (PTN3_HUMAN) | |
| ZASP | Loss-of-function variant of ZASP reduces I Na and modulates the voltage dependence of activation and inactivation | Dilated cardiomyopathy and cardiac noncompaction | Not determined | O75112 (LDB3_HUMAN) | |
Immunofluorescence staining experiments have shown that Na V 1.5 is expressed in distinct membrane compartments, that is, the intercalated discs and the lateral membrane of cardiac cells , , ( Fig. 18.2 ). Our group proposed that the molecular determinants of Na V 1.5 differ between membrane compartments (“pools”). We showed that Na V 1.5 belongs to the dystrophin multiprotein complex based on strong molecular and in vivo evidence, , whereas dystrophin and syntrophin proteins are mainly expressed at the lateral membrane, and not at the intercalated discs of human, rat, or mouse cardiomyocytes ( Fig. 18.3 ). Roden and colleagues investigated a knock-in mouse model harboring the p.D1275N mutation in the gene SCN5A, which codes for Na V 1.5 and is found in patients with dilated cardiomyopathy. They observed a marked reduction in the expression of Na V 1.5 exclusively at the lateral membrane. Makara and coworkers reported that ankyrin-G specifically permitted the targeting of Na V 1.5 to the intercalated discs. A third pool of Na V 1.5 likely exists at the T-tubules, based on functional and morphologic evidence. , , , Milstein and colleagues showed that Na V 1.5 colocalized with SAP97 at the T-tubules. The aforementioned evidence, however, needs to be interpreted with caution because a T-tubular sodium current can be assessed only indirectly, whereas immunofluorescence data , , , lacked fiducial T-tubular markers. Vermij and coworkers could not reproduce the aforementioned functional evidence for a T-tubular Na V 1.5 pool, yet convincingly showed T-tubular Na V 1.5 expression by correlating Na V 1.5 signal with the structural T-tubular marker Bin1 using single-molecule localization microscopy and novel computational methods.
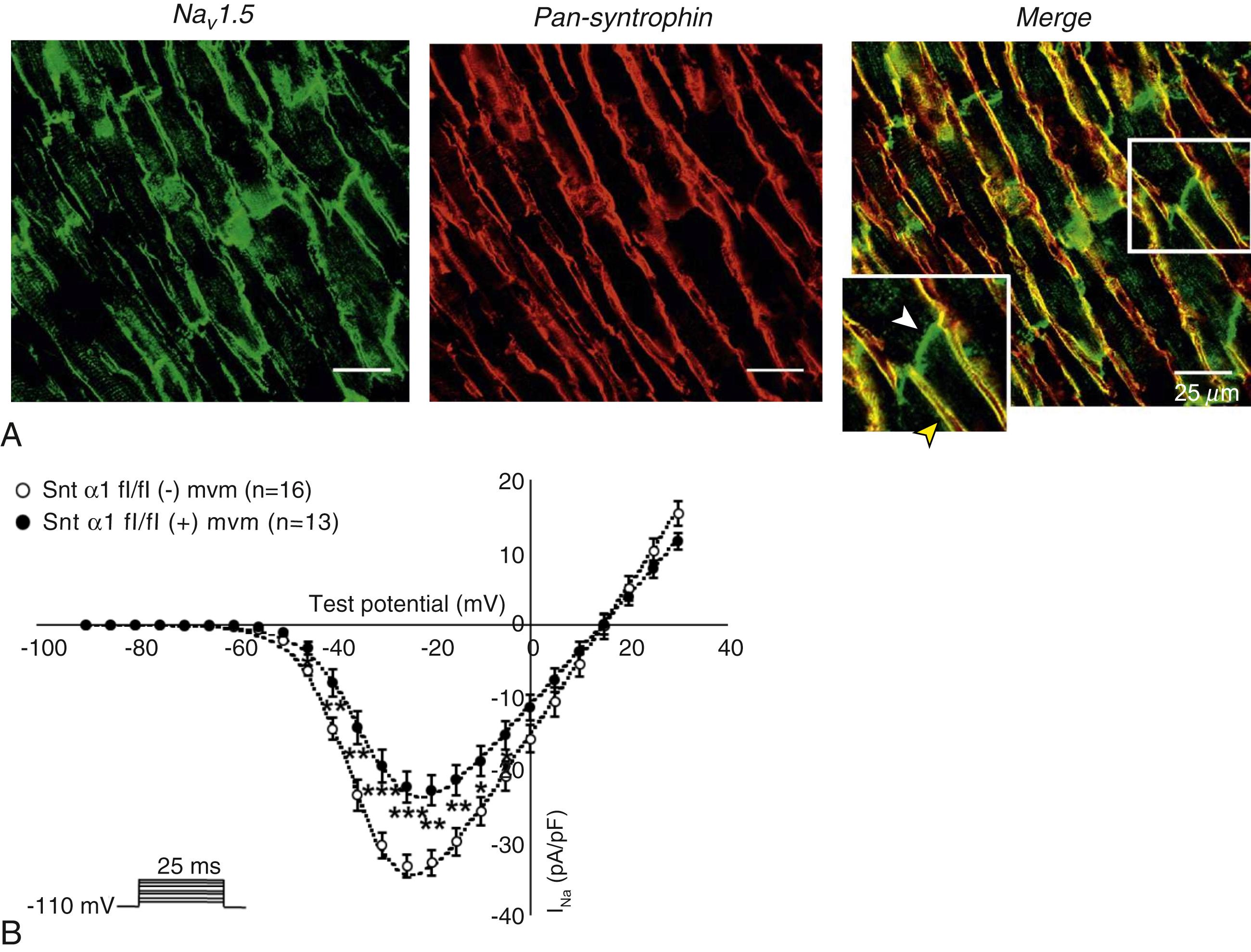
At the intercalated disc, Na V 1.5 depends on the expression of key intercalated disc proteins, such as connexin-43, , plakophilin-2, and desmoglein-2. Superresolution microscopy techniques also demonstrated that Na V 1.5 proteins form distinct clusters approximately 100 nm in diameter and overlapping with N-cadherin at the discs. Similar clusters are also observed in other membrane compartments of cardiac cells. The location and size of Na V 1.5 clusters at the intercalated disc may be crucial for ephaptic coupling, as opposed to classical electrotonic coupling through gap junctions. Although its relevance in vivo remains elusive, ephaptic coupling is defined as electrical field interactions between membranes that are very close together. A depolarized cardiomyocyte may cause a large negative potential in the intercalated disc cleft, which may depolarize the neighboring cardiomyocyte. ,
Thus not only does Na V 1.5 interact with many different partner proteins, but many interacting proteins are location specific. In other words, there is not just one cardiac sodium channel Na V 1.5, but there is a multiplicity of them subject to variable regulatory mechanisms.
This section lists the proteins that interact with Na V 1.5 that have no demonstrable pathologic roles (yet). These interacting proteins were discovered either by protein-protein interaction screens, such as yeast two-hybrid assays, or by proteomic-based protein identification assays. The sites of interaction, often protein-protein interaction domains, were mapped on the sequence of Na V 1.5 as described in Fig. 18.1 . This list follows the chronologic order of published studies.
Protein ubiquitination tags them for degradation but also serves multiple “nondegradative” functions, in particular membrane protein trafficking. Ubiquitin is a small protein of 76 amino acids found in all animal cells. E3 ubiquitin protein ligases mediate the covalent ubiquitin binding to lysine residues of target proteins. Ubiquitinated membrane proteins are internalized and can be targeted for lysosomal or proteasomal degradation, or deubiquitinated by specific proteases and recycled back to the membrane. , The Nedd4-like family of E3 ubiquitin protein ligases regulate many different ion channels. Nedd4-like enzymes specifically bind to target proteins that have consensus domains known as PY motifs with the sequence [L/P]P x Y ( x is any amino acid). , Nedd4 and Nedd4-like enzymes harbor several WW domains that can interact with these PY motifs. In Xenopus oocytes, Nedd4-2-mediated ubiquitination decreases I Na conducted by Na V 1.5. Our group found that the ubiquitin-protein ligase Nedd4-2 directly binds to the PY motif of Na V 1.5 and ubiquitinates the channel. Because the total Na V 1.5 protein level did not decrease upon Nedd4-2 coexpression, we concluded that Nedd4-2 most likely induces increased Na V 1.5 internalization rather than degradation. On the other hand, inhibition of Nedd4-2 by phosphoinositide 3 kinase may cause increase in Na V 1.5 expression at the cell membrane. Inhibition of the proteasome is associated with an increase in I Na and Na V 1.5 expression in neonatal rat cardiomyocytes. Ubiquitinated Na V 1.5 was found in mouse cardiac tissue, further suggesting that ubiquitination regulates Na V membrane turnover or stability in vivo. , Interestingly, the effects of Nedd4-2 on Na V 1.5 partly depend on the intracellular calcium concentration: an increase in intracellular calcium increased Nedd4-2 activity, Nedd4-2-Na V 1.5 interaction, and Na V 1.5 ubiquitination in transfected neonatal rat cardiomyocytes and HEK293 cells. Correspondingly, a rat heart failure model showed increased Nedd4-2 and decreased Na V 1.5 protein levels. Whether these findings correlated with an increase in intracellular calcium was not tested, but cytosolic calcium overload in heart failure is a widely reported phenomenon.
An SCN5A mutation (p.Y1977N) in the Nedd4-2 binding motif constitutes a gain-of-function mutation and is associated with LQTS3. , This mutation abolished Na V 1.5–Nedd4-2 interaction, and prevented Nedd4-2-induced I Na decrease in HEK293 cells and in cardiomyocytes from p.Y1977N knock-in mice; however, these knock-in mice surprisingly show no changes in action potential parameters or persistent I Na . On the other hand, an SCN5A mutation introducing an extra PY motif (p.L1239P; from LL x Y to LP x Y) constitutes a loss-of-function of channel function and is associated with BrS. Na V 1.5-p.L1239P shows increased ubiquitination and degradation in HEK293 cells. Interestingly, the SUMO (small ubiquitin-like modifiers)-conjugating protein UBC9 has been shown to interact with Nedd4-2. UBC9 overexpression increased Na V 1.5 ubiquitination, reducing Na V 1.5 protein expression and I Na . Whether UPC9 only acts on Na V 1.5 indirectly via Nedd4-2 remains unknown. Altogether, these results strongly suggest that the ubiquitin-proteasome system is involved in Na V 1.5 regulation and plays a role in cardiac disease.
The 14-3-3 protein family contains ubiquitous dimeric cytosolic adaptor proteins. The members of this family are involved in many cellular functions, such as the binding and regulation of trafficking of various membrane proteins. Allouis and colleagues performed yeast two-hybrid and coimmunoprecipitation experiments showing that the isoform 14-3-3η interacts with Na V 1.5 at its N-terminal part of the intracellular loop linking domains I to II (see Fig. 18.1 and Table 18.1 ). 14-3-3 and Na V 1.5 colocalize at the intercalated discs of rabbit cardiomyocytes. No influence on the Na V 1.5-mediated peak current was observed when Na V 1.5 and 14-3-3η were coexpressed in CV-1 in Origin carrying SV40 genetic material (COS) cells, suggesting that this protein does not affect Na V 1.5 trafficking. In this expression system, 14-3-3 shifted the inactivation curve toward more negative potentials and delayed recovery from inactivation, illustrating that 14-3-3 proteins can modify the biophysical properties of ion channels. Utrilla and coworkers illustrated the complexity of 14-3-3 regulation as inhibition of 14-3-3 proteins did not modify I Na in Chinese hamster ovary (CHO) cells transfected with Na V 1.5 only but decreased I Na and I K1 when cells were cotransfected with Na V 1.5 and K ir 2.1 without abolishing the Na V 1.5–K ir 2.1 interaction. Interestingly, 14-3-3 is also proposed to mediate Na V 1.5 dimerization and coupled gating (i.e., biophysical coupling of two Na V 1.5 α-subunits). , , Because different isoforms of 14-3-3 proteins are expressed in cardiac cells, , their exact roles in normal cardiac function and their implications in disease states require further investigation.
Calcium/calmodulin-dependent protein kinase II (CaMKII) is a serine/threonine protein kinase expressed in many cell types, and it transduces intracellular calcium increases into the phosphorylation of target proteins, including cardiac ion channels. , CaMKIIδc is the predominant cardiac isoform upregulated in human and animal heart failure models. , Na V 1.5 colocalizes and coimmunoprecipitates with CaMKIIδc in transfected HEK293 cells, rabbit and rat cardiomyocytes, and mouse heart lysates. , , Ashpole and colleagues have shown that CaMKIIδc interacts with the first intracellular loop of Na V 1.5 (see Fig. 18.1 ) and that residues Ser516 and Thr594 may be phosphorylated by this kinase. Hund and associates found that Ser571 within the same Na V 1.5 intracellular loop was also a target of CaMKII isoforms α, β, γ, and δ in a β IV -spectrin-dependent manner. β IV -spectrin interacts with ankyrin-G at the intercalated discs, suggesting Ser571 phosphorylation may be specific to the intercalated disc. Overexpression of CaMKIIδc in rabbit cardiomyocytes and transgenic mice induced a calcium-dependent hyperpolarizing shift of the steady-state inactivation curve, slowed the recovery from inactivation, and increased the persistent/late I Na . Similar effects on I Na are observed with certain congenital LQTS type 3 mutations in SCN5A. In mice, transgenic overexpression of CaMKIIδc leads to chronic heart failure and ventricular tachycardia episodes. In dog ventricular myocytes, increased CaMKII activity increases the persistent I Na . Whether these observations are the direct consequence of the Na V 1.5 biophysical alterations or related to other heart failure mechanisms remains unclear. In atrial samples from atrial fibrillation patients, phospho-Na V 1.5 at Ser571 and autophosphorylated CaMKII were increased, correlating with the increased persistent I Na and calcium handling defects in murine atria expressing phosphomimetic Na V 1.5-S571E. Similarly, in atrial samples from patients with sleep-disordered breathing, which is often associated with atrial arrhythmias, activated CaMKII levels, persistent I Na , and phospho-Na V 1.5 at Ser571 were increased, whereas peak I Na was decreased because of a shift in steady-state inactivation. Taken together, CaMKII is an important component in the pathogenesis of arrhythmias and a potential drug target. ,
El Rafaey and colleagues recently described protein phosphatase 2A (PP2A) as a negative regulator of the Na V 1.5/CaMKII axis. PP2A is a serine-threonine phosphatase with a catalytic, scaffolding, and regulatory subunit. Cardiomyocytes from mice lacking the B56α regulatory subunit show increased PP2A activity, less Na V 1.5 phosphorylation at Ser571, and less persistent sodium current on isoproterenol treatment than wild-type cells. PP2A colocalizes with Na V 1.5 at the intercalated disc, and interacts with Na V 1.5, probably mediated by ankyrin-G, as shown in coimmunoprecipitation experiments. Thus PP2A may play an important role in modulating CaMKII-induced Na V 1.5 phosphorylation and the persistent sodium current.
Ion channels are also regulated by phosphorylation of tyrosine residues, a process that depends both on the phosphorylation activity of tyrosine protein kinases and the dephosphorylation by phosphatases. Overexpression of protein tyrosine kinase Fyn in HEK293 cells was shown to alter several biophysical properties of Na V 1.5 via the phosphorylation of Tyr1495. This residue is close to the Ile-Phe-Met (IFM) cluster in the intracellular loop linking domains III–IV (see Fig. 18.1 ), which mediates the rapid inactivation of voltage-gated sodium channels. Coexpressing Fyn with Na V 1.5 in HEK293 cells shifted the I Na steady-state inactivation curve toward depolarized potentials and accelerated the recovery from inactivation. The interaction site of Fyn with Na V 1.5 remains to be mapped.
Our group reported that the protein tyrosine phosphatase PTPH1 interacts with the PDZ-domain-binding motif of Na V 1.5. PTPH1 coexpression in HEK293 cells shifted the availability curve of wild-type Na V 1.5 toward hyperpolarized potentials. This effect was abolished when the PDZ-domain-binding motif of the Na V 1.5 C-terminus was removed by an early stop mutation. These results suggest that tyrosine phosphorylation of Na V 1.5 modulates the stability of the inactivated state. The fact that several proteins (e.g., PTPH1, syntrophin proteins, and SAP97) interact with the same binding domain supports the model that different Na V 1.5 multiprotein complexes coexist within cardiac cells.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here