Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Major advances have been made over the last 3 decades that have defined the genetic basis of many medical diseases. There are now more than 40 different cardiovascular diseases directly caused by variants in genes that encode cardiac proteins. These cardiovascular diseases include inherited cardiomyopathies, inherited arrhythmia syndromes, metabolic disorders, familial aortopathies, and congenital heart diseases. Identification of the genetic causes of cardiovascular disease allows early detection of at-risk asymptomatic family members and, in some cases, helps guide therapies and inform prognosis. This chapter provides an overview of the current knowledge related to the role of genetic testing in cardiac diseases, with a specific focus on arrhythmogenic diseases.
With the completion of the human genome sequence and subsequent advances in genetic technologies, we now have a clearer picture of our genetic makeup and how variations in our genome can lead to cardiovascular disease.
The human genome is made up of approximately 3.2 billion base pairs of DNA (composed of four bases: adenine, thymine, guanine, and cytosine), accounting for approximately 22,000 genes. Each gene is defined as a molecular unit that can encode both RNA and protein sequences, which represent functional units in the human body. In cardiovascular disease, these genes can encode both RNA and proteins, and variants in these genes can lead to a disease phenotype. Functionally, disease genes leading to cardiovascular disease encode a range of proteins, including those associated with the sarcomere, ion channels, cytoskeletal structures, and in embryonic heart development.
Variations, or changes, can occur in the DNA sequence of humans. These variants can be broadly defined as single nucleotide variants (SNVs). SNVs usually occur with a measurable frequency (>0.5% allelic frequency) among a particular ethnic population. SNVs can then be subdivided into those that alter the protein sequence (nonsynonymous) and those where the protein sequence is unaltered (synonymous). Rare variants are seen in everyone, so determining which can cause disease is a significant challenge. The American College of Medical Genetics and Genomics (ACMG) have developed stringent classification criteria for assigning causation to rare variants.
Many types of disease-causing variants exist. The vast majority in cardiovascular disease are missense variants, in which a single base pair change results in the change (or replacement) of one amino acid. Other variants may cause more significant disruptions to the encoded protein, leading to a shift in the reading frame and/or downstream premature protein truncation. This can lead to a major change in the protein sequence or a loss of amino acids resulting in a shortened protein. The latter variants are often caused by insertions or deletions of nucleic acids in the coding region. The terms pathogenic or likely pathogenic imply there is a level of confidence to pursue cascade genetic testing of asymptomatic relatives ( Table 69.1 ).
| Possible Outcome | Consequences for the Proband | Consequences for the Family |
|---|---|---|
| No variants of potential clinical importance identified (benign) | An indeterminate gene result does not exclude a cardiac genetic disease, but reassessment of the phenotype should be considered. | Cascade genetic testing cannot be offered to the family; at-risk relatives are advised to be clinically assessed according to current guidelines. |
| Variant of uncertain significance identified | Efforts to delineate pathogenicity of the variant are required, including cosegregation studies involving phenotyped family members. | Although pathogenicity of a variant is under question, it cannot be used to inform clinical management of family members; cascade genetic testing cannot be offered; at-risk relatives are advised to be clinically assessed according to guidelines. |
| Pathogenic variant identified (pathogenic or likely pathogenic) | Confirm clinical diagnosis, limited therapeutic and prognostic application except in familial long QT syndrome. | Cascade genetic testing of asymptomatic family members is available after genetic counseling. |
| Multiple pathogenic variants identified | Confirm clinical diagnosis and potentially explain a more severe clinical phenotype. | Complex inheritance risk to first-degree relatives must be discussed; cascade genetic testing of asymptomatic family members is available after genetic counseling. |
| Incidental or secondary pathogenic variant identified | Action regarding incidental or secondary findings must be discussed with the proband pretest. | Genetic counseling to determine clinical and genetic impact on family members is available. |
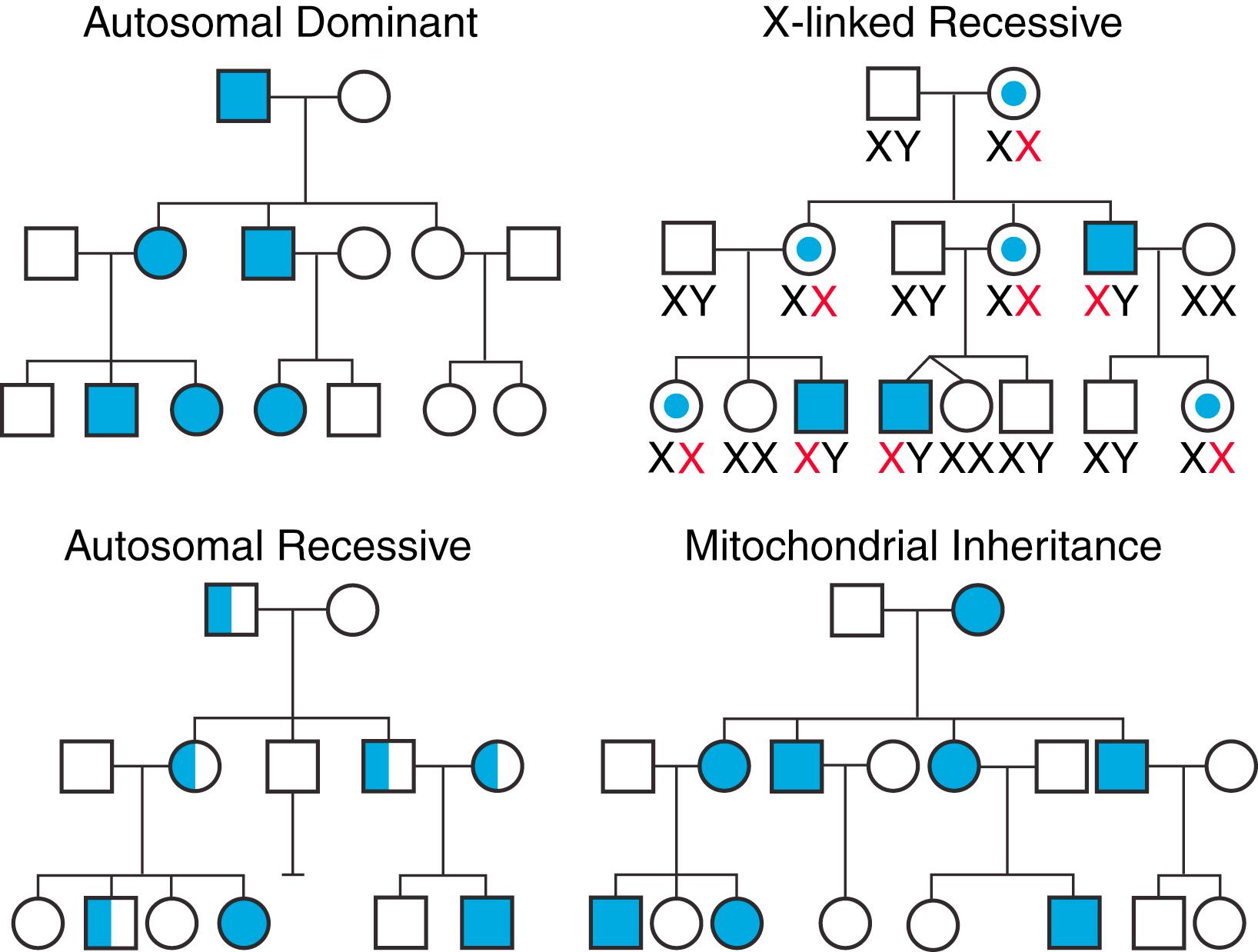
The four major modes of inheritance are summarized in Fig. 69.1 . In the majority of cardiovascular genetic diseases inheritance is autosomal dominant, whereby the disease can be expressed when the variant is present in just one allele. As a result, the chance of passing on the gene variant from parent to offspring is 50%. Most primary arrhythmogenic diseases and inherited cardiomyopathies are inherited in this fashion. The other three modes of inheritance in Fig. 69.1 are significantly less common. Autosomal recessive inheritance requires the person to inherit the variant on both alleles for disease to develop (i.e., one variant inherited from each parent). The chance of passing on the gene variant from both parents to offspring is 25%. The Jervell-Lange-Nielsen form of familial long QT syndrome (LQTS) is inherited in an autosomal recessive fashion. X-linked inheritance refers to the situation in which the gene variant is located on the X chromosome. As such, males develop the phenotype, whereas females are most often asymptomatic gene variant carriers. Rare forms of dilated cardiomyopathy (DCM) have been shown to have an X-linked pattern of inheritance. Mitochondrial inheritance is a non-mendelian pattern in which transmission of disease is exclusively via females and involves inheritance of mutant mitochondrial DNA to offspring. Although uncommon, this form of inheritance is often seen in mitochondrial diseases that can clinically manifest with a hypertrophic cardiomyopathy (HCM) phenotype (or genocopy).
Genetic testing is not a simple blood test. Many considerations arise with every family. A complete cardiogenetic evaluation is required, which includes a thorough cardiac investigation and a clear diagnosis in the proband, the need for genetic counseling to ensure that the family understands the possible outcomes of genetic testing, and taking a detailed family history to get a sense of disease penetrance and patterns of disease. ,
The cornerstone of the utility of genetic testing is accurately defining the clinical phenotype both in the individual patient and the family. The highest yields from genetic testing are often based on patient cohorts with confirmed disease. For example, in clinical HCM, careful attention to the family history, clinical symptoms, and defining the extent, distribution, and severity of hypertrophy are all considered essential in clinically distinguishing HCM from other HCM mimickers (or genocopies), such as Fabry disease or glycogen storage diseases, which have different genetic etiologies.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here