Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
We sincerely thank Dr. Ali Ravanpay for his work on the previous edition of this chapter, which was critical for the updates provided in this new version.
This chapter includes an accompanying lecture presentation that has been prepared by the authors: ![]() .
.
IDH1 mutations significantly influence the survival of patients with gliomas.
Single-cell RNA sequencing demonstrates significant intratumoral heterogeneity within glioblastomas.
Analysis of primary and recurrent glioblastoma samples demonstrated recurrent tumor clones diverged from a common ancestor more than a decade before diagnosis.
Patient-specific glioma organoids with unique genetic alterations provide a representative and robust in vitro platform for drug screens on both the tumor and microenvironment.
Immunocompetent and bilateral glioma mouse models are providing an in vivo platform for assessing the impact of the immune system on treated and untreated tumors with distinct genomic alterations within the brain.
Gliomas are CNS tumors believed to be derived from astrocytes or their precursors. , The World Health Organization (WHO) classifies gliomas based on histopathologic criteria including predominant cell type, vascularity, mitosis, and necrosis. Grade IV gliomas such as glioblastoma multiforme (GBM) represent the highest and deadliest grade of disease, with a median survival of 14 months, even with aggressive surgical resection and adjuvant chemotherapy and radiotherapy. Unfortunately, the only significant advancement in survival (approximately 2 months) was made when Stupp and colleagues found that temozolomide (TMZ) combined with radiotherapy improved the survival of patients with GBM compared with patients receiving radiotherapy alone. The difficulties in obtaining improvements in GBM patient survival are likely caused in part by disease resistance and recurrence mediated by intratumoral heterogeneity.
Intratumoral heterogeneity represents diversity within the tumor, with some cells driven by distinct molecular alterations and mutations. , This diversity makes therapeutic intervention difficult because different molecular alterations confer differential susceptibility to chemotherapeutic agents. Before the current era of technologic advancement, it was thought that GBMs represented one disease entity. However, molecular and genomic analyses demonstrate that GBMs exist in multiple molecular subtypes, and each tumor is composed of multiple clones of cells with different phenotypic characteristics. , ,
Ultimately, therapeutic success against GBMs will only arise following delineation of the key molecular drivers of the disease and the discovery of drugs that effectively target cells with these molecular alterations. Although it is unclear if this targeted therapy strategy (or a combination of therapies) will be effective, it may provide an avenue for making conventional therapy more effective by minimizing diversity and heterogeneity within tumors. In this chapter, we discuss ongoing strategies aimed at elucidating the molecular signatures of gliomas, especially GBMs. We provide an overview of basic, preclinical and clinical techniques for characterizing cells within these tumors. We also show how these techniques are providing clues for the development of therapeutic targets for gliomas.
The WHO Classification of Tumors of the Central Nervous System was recently updated to incorporate advances in the molecular profiling of CNS tumors. The most recent iteration includes both genomic and epigenomic alterations that define specific subtypes of adult and pediatric brain tumors with significant clinical consequences (therapy and prognosis). In pediatric gliomas, there is now a distinct H3K27-mutant diffuse midline glioma that is associated with a poor prognosis but provides a therapeutic target in these patients. For adult gliomas, the most significant updates include the recognition of IDH1 wild-type versus IDH1 -mutant glioblastomas as distinct entities with prognostic utility (IDH1-mutant gliomas have a more favorable prognosis). Now there are efforts to target IDH1 therapeutically or stratify responses in clinical trials based on IDH1 status to better delineate which patients are optimal candidates for specific drugs already in clinical use. In this chapter, we explore the detection of these and other molecular signatures that could guide future classification systems for gliomas with therapeutic implications.
The past three decades have witnessed unprecedented progress in the development of groundbreaking techniques in molecular and cellular genetics that enable the study of DNA. Oncogenesis occurs either as a result of transmitted preexisting DNA mutations or DNA changes occurring over time in response to a reciprocal and dynamic crosstalk between the cellular microenvironment and the cellular DNA content. The study of cancer, which is ultimately a disease of abnormal DNA, relies on tools to identify, isolate, and characterize these abnormalities. In this section, we introduce some of the earliest techniques in isolating, amplifying, and studying DNA changes associated with cancer development. Then we focus on the more recent complex, large-scale techniques allowing comparative studies between candidate cancer cells and normal cells.
The first step in characterizing aberrant tumor DNA is the ability to amplify small amounts of DNA found in tumor cells without introducing new errors, such that enough DNA substrate material is generated for further molecular cytogenetic studies. Polymerase chain reaction (PCR), discussed further in the next section, allows the large-scale replication of DNA for additional study. After amplification of target DNA sequences, the simplest next step is DNA sequencing, allowing a readout of the genetic code and future comparison among sequences obtained from diseased host cells and normal controls. In this way, candidate mutations resulting in more important changes in cellular behavior have been identified. After isolating and amplifying target DNA sequences, researchers can then introduce (or remove) specific DNA sequences in cells and animals to understand the function of these sequences. Such experiments require introduction of PCR-generated amplified DNA into expression vectors using continually improving techniques in molecular biology and DNA expression construct synthesis. To study the functional consequences of mutations identified by DNA sequencing techniques, target DNA sequences must be introduced into host cells in culture by using either the nonviral transfection process or virus-based gene transfer. Beyond these simple methods, large-scale genome DNA and RNA techniques are allowing researchers to study vast sequences of genomic DNA and evaluate the relative levels of expression of DNA or RNA transcripts in pathologic conditions as compared with normal. Here we describe some of the key techniques currently in use and in development for the study of cancer biology.
PCR has revolutionized the applicability of gene-based technologies to clinical medicine. It permits the identification and amplification of DNA sequences from any source, as long as part of the sequence is known. Therefore, any known DNA sequence can be detected in any sample containing genetic material, including tumors.
Initially, two oligonucleotide primers (short pieces of synthetic DNA with sequences designed to anneal, or “stick,” to regions flanking the DNA sequences to be amplified) are designed and synthesized chemically. DNA containing the target sequence to be amplified is mixed together with oligonucleotide primers, free nucleotides, and heat-stable DNA polymerase enzymes. Each cycle of PCR requires heating the mixture to melt double-stranded DNA into two single strands, cooling to anneal primers to the template DNA to be amplified, synthesizing a new complementary strand from free nucleotides, and, finally, melting and annealing to separate new strands of DNA from old strands. Each cycle results in doubling of the initial template quantity. After 10 cycles of PCR, the original template DNA is amplified approximately 100 times, and after 20 cycles, approximately 1 million times ( Fig. 136.1 ). Once a PCR product is separated on a gel, DNA fragments of expected size are collected, and the nucleotide sequence is checked to verify amplification of the correct target.
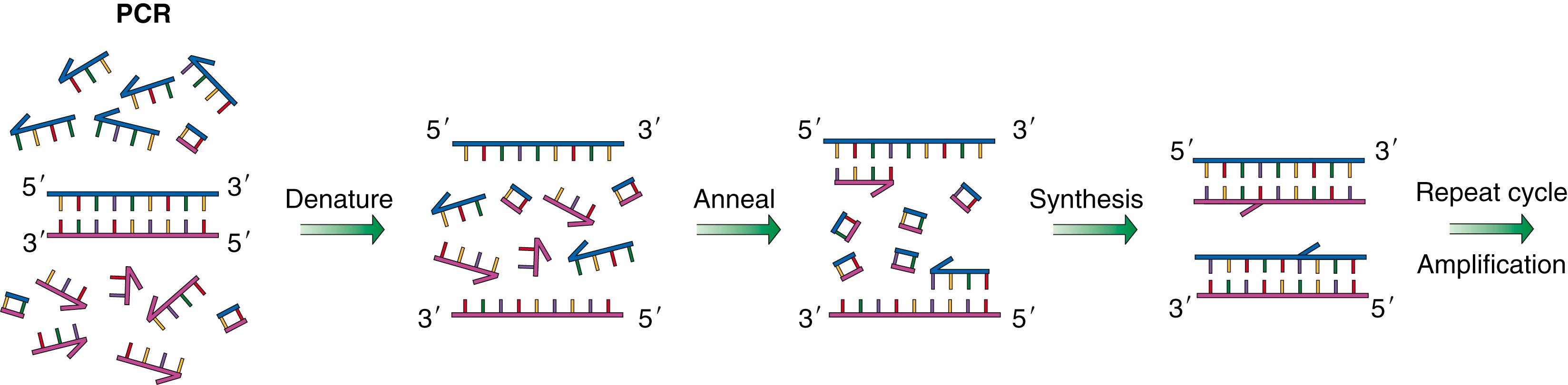
PCR can be used to detect or quantify particular DNA sequences in a sample, for example, to detect the presence of viral sequences or to determine whether a tumor sample contains known sequences or a known genetic alteration. PCR products can also be used to generate purified DNA of known sequences for use in the construction of novel DNA molecules. Perhaps most important, PCR is now used routinely to detect new mutations in amplified or deleted regions of tumor DNA and new mutations in known or putative tumor suppressor genes and oncogenes.
In its simplest form, DNA sequencing uses DNA substrate, frequently isolated from PCR reactions (or occasionally from other DNA purification approaches), to study the exact order of the four nucleotides in a stretch of DNA. This technique has undergone tremendous advances since its conception in the early 1970s. Initially, and as late as the early 1990s, researchers used two-dimensional chromatography and radiolabeled nucleotides to sequence DNA. Since the advent of fluorescence-based techniques, DNA sequencing has become automated, allowing simpler, faster, and more accurate sequencing of significantly larger stretches of DNA. This simple sequencing technique has served as the basis for more complicated approaches, discussed later in this chapter, to look at more details of DNA sequence characteristics.
Sequencing an organism’s whole genome can be accomplished at low cost nowadays. Several techniques exist for accomplishing whole-genome DNA sequencing : one is the shotgun approach ( Fig. 136.2 ). This technique involves cutting genomic DNA into 150-Mb segments using restriction enzymes that recognize specific sequences on DNA. These pieces of DNA are then amplified by cloning into a plasmid or a bacterial artificial chromosome (BAC) vector and introducing the construct into the Escherichia coli bacterium . Following selection for E. coli bearing the vectors, the plasmid is then sequenced using primer sites outside of the sequence to be analyzed. Following multiple sequencing runs (depending on the depth of sequencing required, as discussed later), identical sequences begin to overlap. When these overlapping sequences are aligned, the complete sequence of the organism can be assembled. To obviate the amplification steps in this approach, which may lead to the introduction of errors, other researchers have developed an in vitro–only technique wherein DNA fragments, following digestion, are amplified on primers attached to a solid surface. Other researchers use fluorescent nucleotides during this event, and laser excitation is used to detect the addition of individual nucleotides to the growing DNA strand, hence generating a sequence of the parent DNA strand and avoiding the need for amplification.
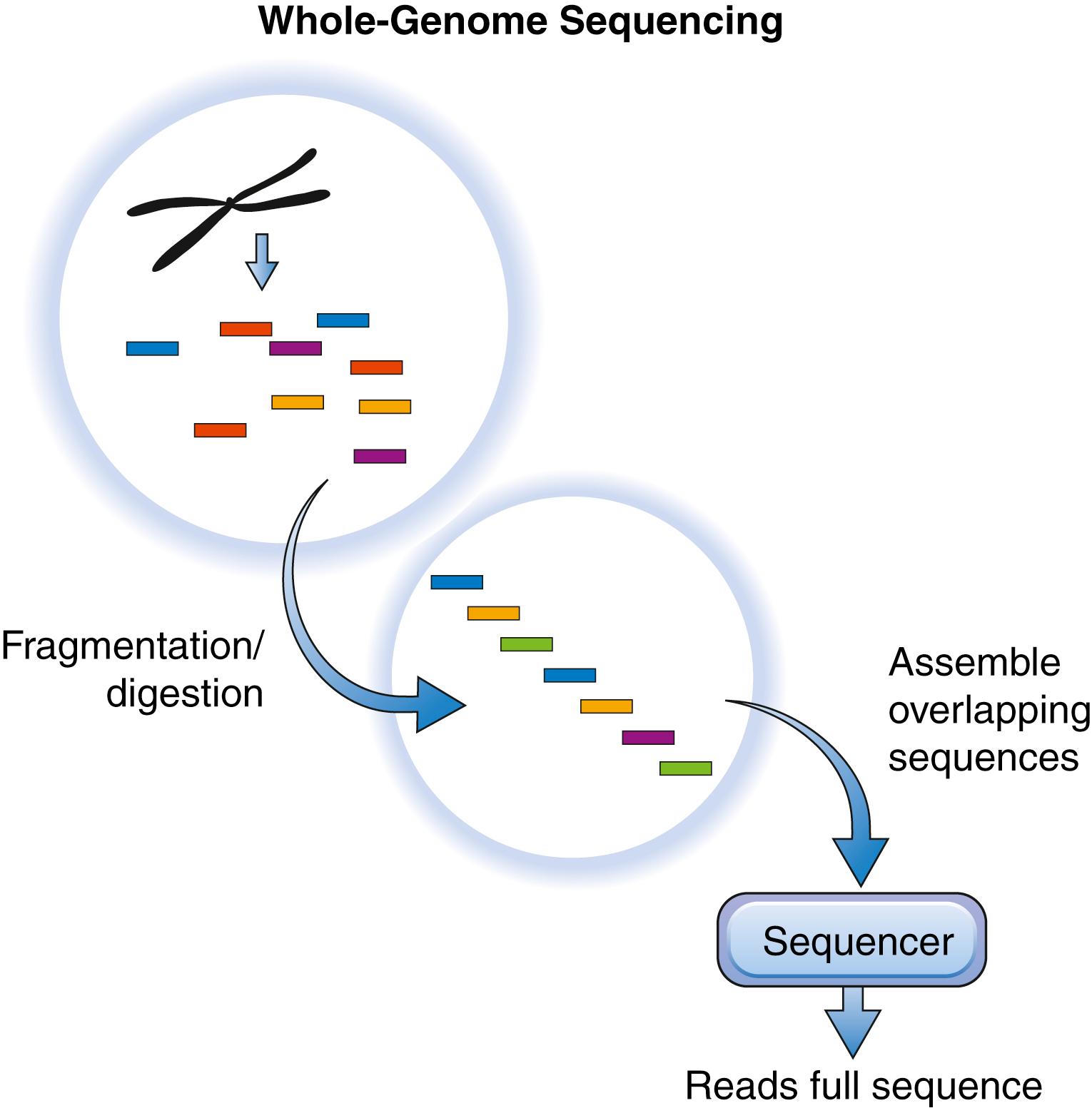
To improve the speed and accuracy of DNA sequencing, new technologies that parallel the sequencing process just described by analyzing millions of sequences concurrently (next-generation sequencing) have been developed over the past decade. Some of these technologies allow for sequencing of a large number of DNA strands while they are being synthesized (“sequencing by synthesis”). The unique feature of the next-generation sequencers is that the process does not require bacterial amplification. Instead, most of the sequencing occurs on solid platforms in vitro with primers already annealed or on adaptor molecules that bind the parent DNA in preparation for PCR-based sequence amplification. Some of the techniques already in development utilize mass spectrometry and microfluidics to allow for simultaneous sequencing of even more DNA strands, with accuracies reaching 98% to 99%.
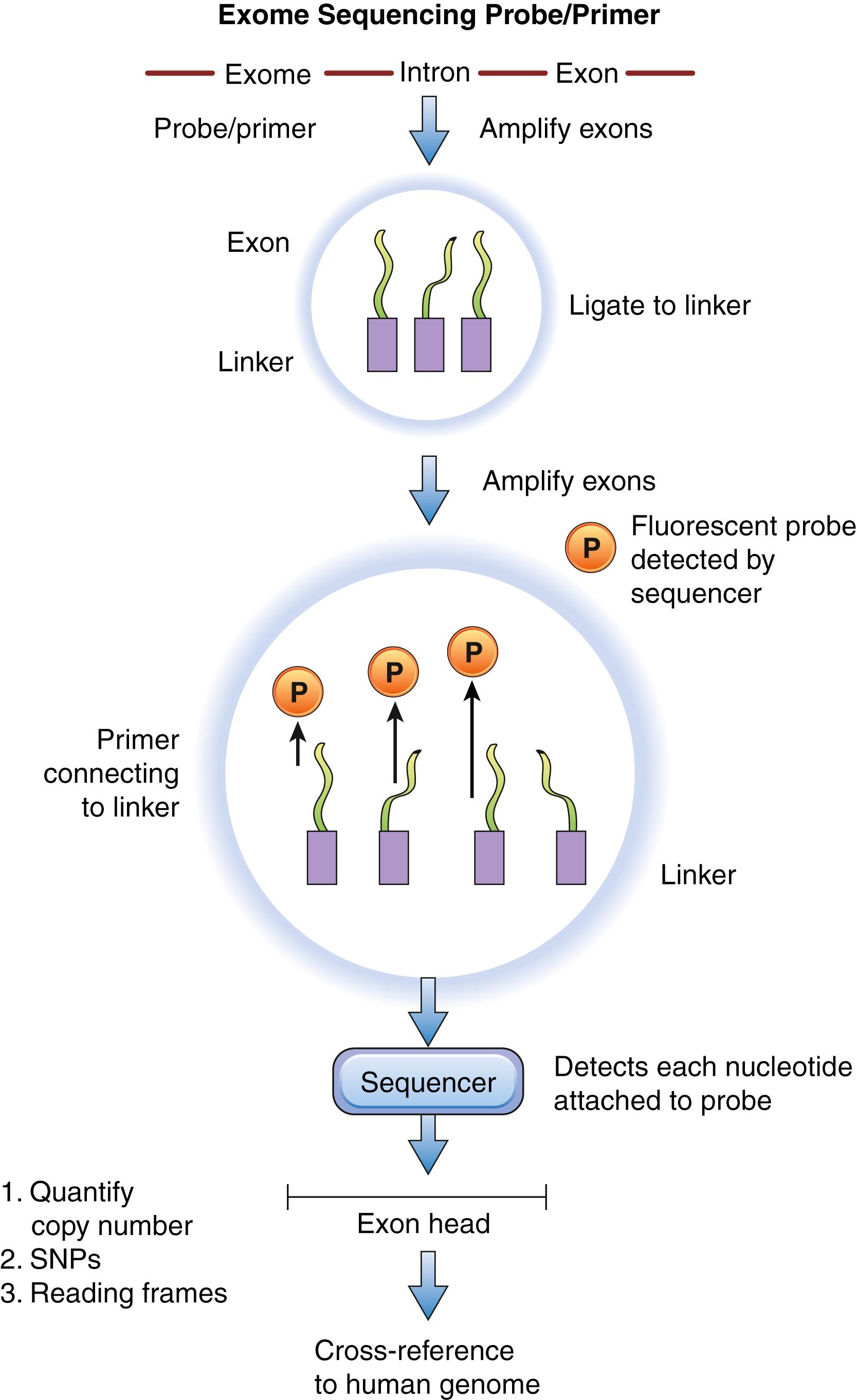
The human exome represents less than 2% of the human genome but contains approximately 85% of known disease-causing variants. , Sequencing the exome only is said to be a cost-effective alternative to sequencing the whole genome because it bypasses the need to sequence introns. The first step of exome sequencing or capture involves selective target enrichment of coding regions of DNA before sequencing. Some techniques utilize PCR to amplify exons; the disadvantage of this technique is that amplification of large DNA regions utilizes a significant amount of template DNA and hence increases cost. An alternative is the molecular inversion probe, which utilizes small oligonucleotides that are flanked by target-specific ends. These probes bind to the flanks of exomes and are circularized following amplification of the exome between the target ends. After removal of the probes that did not circularize, the sequence of exomes can be generated by massive parallel sequencing. Other researchers have developed techniques that utilize beads attached to probes complementary to specific exons. Once the genomic DNA is fragmented, it is mixed with the bead-probe target. The mixture is washed to remove excess material, and the beads are pulled down out of the solution and separated from the parent DNA so as to be sequenced ( Fig. 136.3 ). Ultimately, this technique avoids amplification of the DNA strand via PCR, hence saving time and cost.
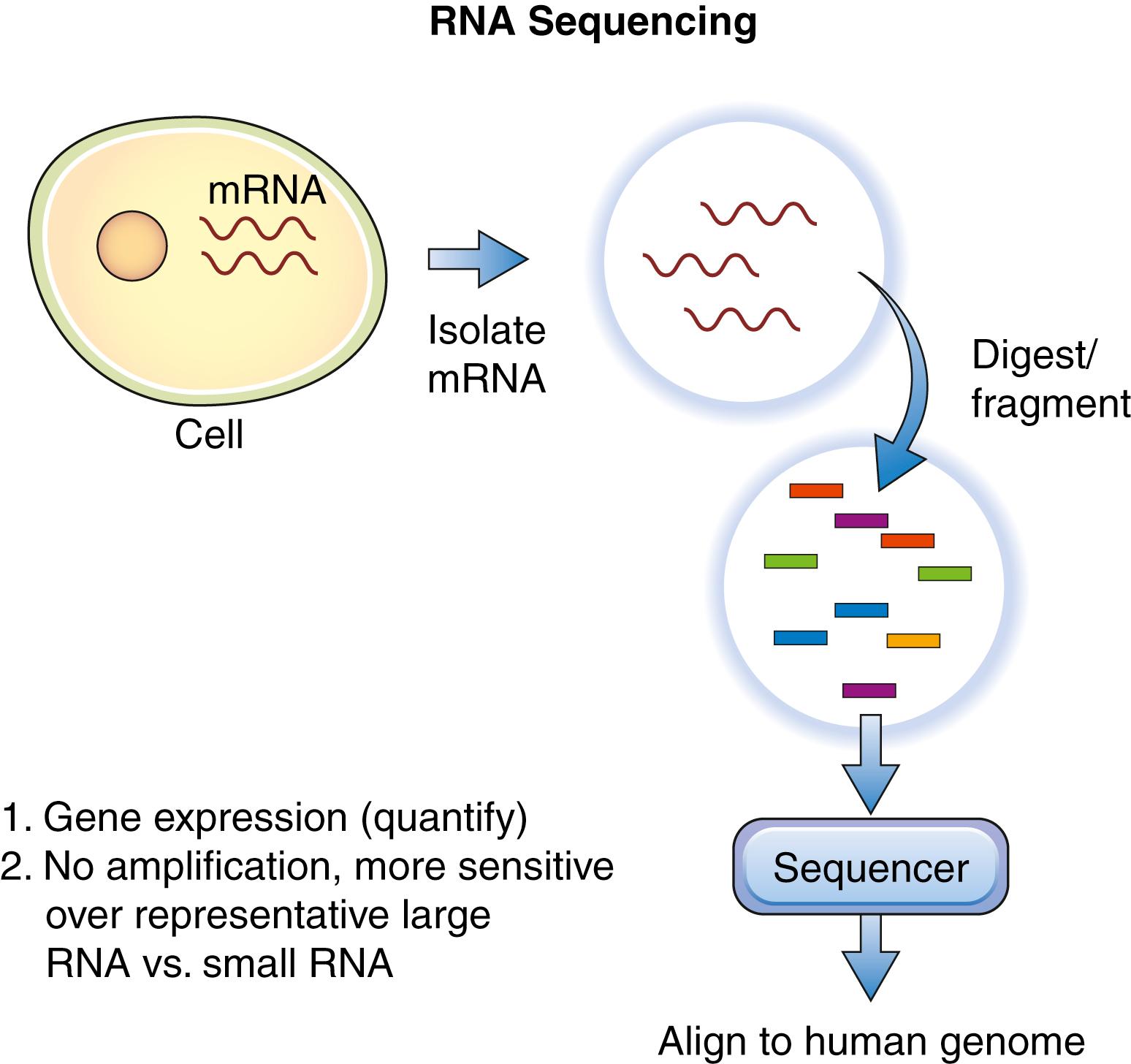
RNA sequencing uses the next-generation sequencing techniques mentioned previously for DNA to provide a glimpse of the relative predominance of RNA at a specific moment in time. , Here the first step following RNA isolation is separation of coding from noncoding RNA. This is accomplished by taking advantage of the fact that coding RNA possesses a poly(A) tail, hence poly(T) oligonucleotide bound to beads can be used to selectively separate coding RNA from noncoding RNA. This technique is particularly useful for the detection of fusion gene products (linked to specific cancers), posttranscriptional alterations, and single-nucleotide variations. Next, the RNA is reverse-transcribed by reverse transcriptase into complementary DNA (cDNA) before sequencing ( Fig. 136.4 ). To minimize errors generated during cDNA synthesis, some researchers have developed platforms that allow for direct RNA sequencing. This can be accomplished de novo (without a genomic reference) or with a genomic reference as a guide. Both DNA and RNA sequencing techniques are still in development, and several years may be required before they are used in the clinical setting.
Given the significant intratumoral heterogeneity within gliomas, especially GBMs, capturing the diversity at the individual cellular level is imperative for finding therapies that eradicate tumor- initiating clones within the tumor and determining how tumors respond to treatment at the clonal level. Clonal evolution during treatment is discussed in the “Genomics Alteration–Driven Clonal Evolution During Treatment” section later in this chapter.
For single-cell RNA sequencing, freshly isolated or fresh-frozen bulk tumors are dissociated into individual cells. Nontumor cells are sequentially removed from the heterogenous mixture. Red blood cells are lysed and centrifuged out of the mix. Immune cells are depleted by fluorescence-activated cell sorting (FACS), which utilizes CD45-immune cell surface markers to sort and select CD45 + -immune cells out of the mix, leaving non–CD45-expressing normal and tumor cells.
Next, individual cells undergo RNA sequencing using the technique described in the “RNA Sequencing” section earlier in this chapter, with most kits allowing for cell lysis, RNA isolation, reverse transcription, and cDNA amplification from small amounts of starting materials such as an individual cell. Finally, to distinguish normal cells from tumor cells during analysis, gene- expression levels from each cell are used to infer copy number variations (CNVs) or copy number alterations (CNAs) within each cell after comparing the data with bulk RNA sequencing data from normal human brain tissue.
In the future, this technique will hopefully allow for specific clones of tumor cells or immune cells that are central to disease progression, which could be targeted by drugs.
Histones are believed to be DNA packaging proteins; however, it is becoming clear that they also affect gene expression depending on specific modifications such as methylation or acetylation. , Specific histone modifications such as histone 3 lysine 27 trimethylation (H3K27me3) have been shown to be associated with repression of adjacent gene expression. Alternatively, histone 3 lysine 4 trimethylation (H3K4me3) is associated with activation of gene expression. In GBMs, proteins that mediate these histone modifications, such as the polycomb group proteins, are frequently deregulated, resulting in aberrantly placed histone modifications. Whole-genome histone modification profiling has demonstrated unique signatures of histone modifications associated with stem cells and cancer. , Therapeutically, it is proposed that targeting these histone modifications, either by enhancing the proapoptotic associated markers or by inhibiting modifications that inhibit tumor suppressor genes, may slow the progression of cancer.
In GBMs, several agents targeting the repressive H3K27me3 modification are currently in development. 3-Deazaneplanocin (DZNep) has been shown to block H3K27 methylation and inhibit proliferation of GBM cells in vitro. Although DZNep is still in the preclinical phase of trials, its discovery suggests that targeting GBMs with histone modification inhibitors may represent another therapeutic avenue for suppressing tumor progression.
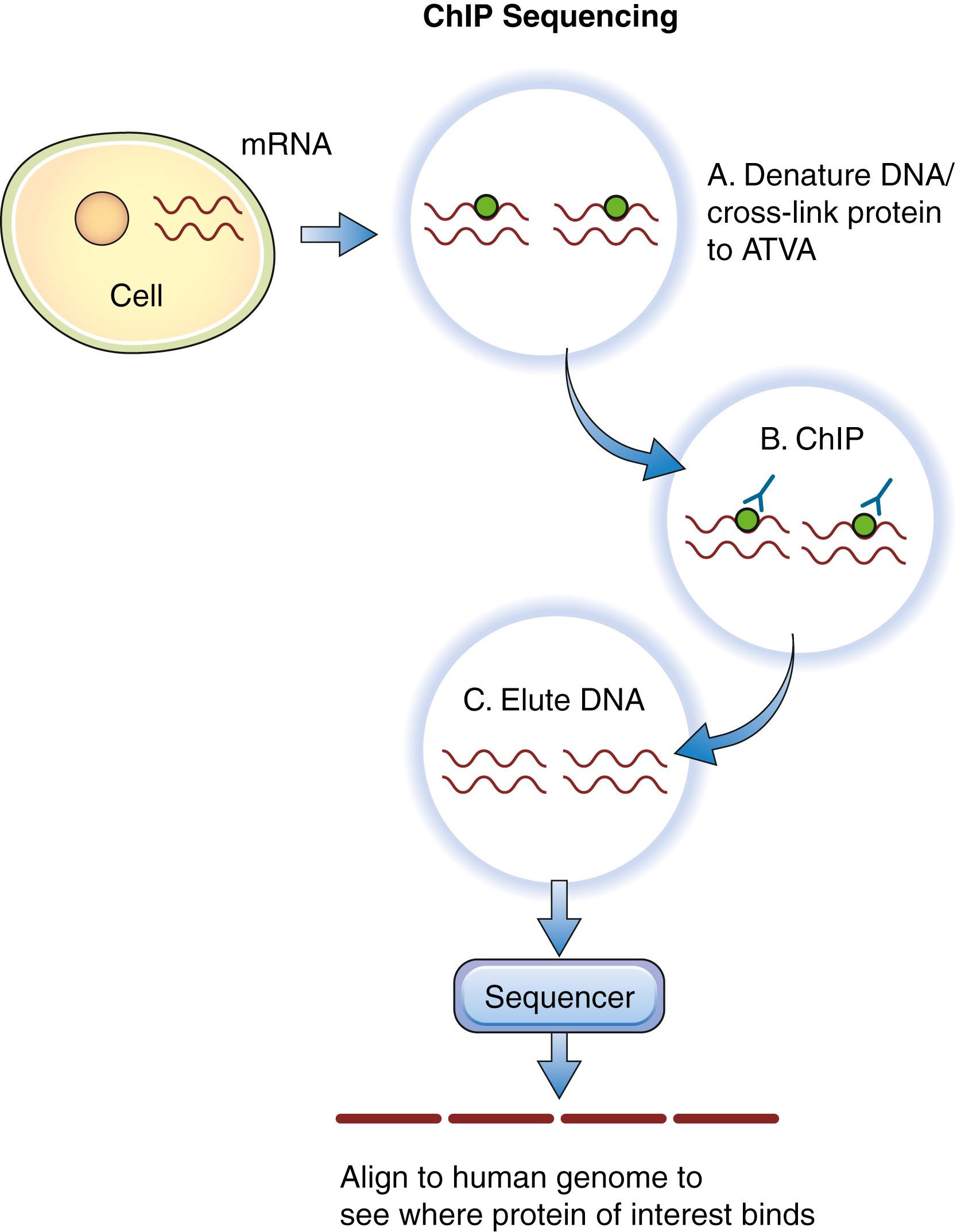
To assess histone modifications at the whole-genome level, antibodies to the specific histone modification of interest are incubated with formaldehyde–cross-linked DNA samples. The cross-linking procedure is performed to maintain proteins that are bound to the DNA in the bound state. The antibodies with histones and associated DNA are pulled down from the mixture. Next, the antibodies, histones, and DNA are denatured and dissociated using chromatin immunoprecipitation sequencing, with the DNA eluted from the mixture given that this sequence represents the target. These individual DNA sequences are then sequenced, and the reads are cross-referenced to the human DNA sequence database ( Fig. 136.5 ). The genes corresponding to these DNA sequences are therefore believed to be associated with the histones targeted in the initial steps of the protocol. Ultimately, when this procedure is performed across multiple tumor samples, unique histone modification signatures at specific genes can be used as a prognostic tool for therapy.
Cancer, especially in its most malignant forms, progresses by accumulating multiple errors in the genomes of affected cells. Many of the genetic errors result in malfunction of the DNA proofreading mechanisms, which normally function to repair damaged DNA. As a result, the entire genome of an affected cell becomes prone to error, resulting in global genomic instability. A consequence of genomic instability is susceptibility to single- and double-stranded DNA breaks. At the chromosomal level, these breaks can result in very large losses, gains, or translocations of chromosomal regions. These chromosomal areas encode genes or regulatory elements of genes involved in cancer initiation and progression, and identification of these abnormal chromosomal regions is important for a general understanding of cancer biology. As a consequence, cytogenetics, which encompasses the study of chromosomal structure, has become a central part of cancer biology.
Before the advent of molecular cytogenetic techniques, the primary method for studying chromosomal aberrations was direct visualization of chromosomes during the metaphase period of the cell cycle. Chromosome condensation during this time allows the highly compact metaphase chromosomes to be directly visualized with light microscopy. Chromosome spreads—metaphase or interphase chromosomes placed on glass slides—can distinguish unique properties of individual chromosomes and allow chromosome regions to be identified. Looking at chromosome spreads can therefore be used to readily identify aberrant chromosome structures or numbers, such as those observed in Kleinfelter and Down syndromes.
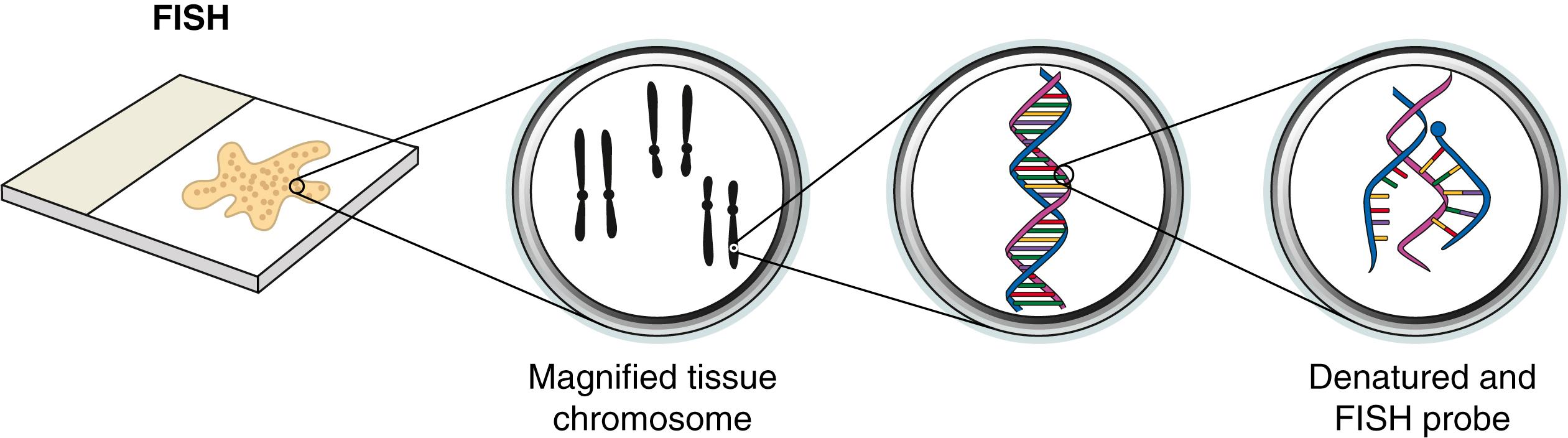
Molecular cytogenetics is an amalgamation of molecular biology and the conventional cytogenetic methods described earlier. The prototypical molecular cytogenetic technique, fluorescence in situ hybridization (FISH), is currently widely used for identification of chromosomal aberrations in cancer genomes. FISH can readily identify large chromosomal abnormalities such as rearrangements and deletions. FISH requires the use of a synthetic oligonucleotide probe containing fluorophores that correspond to a known region of the genome. The probe is then hybridized to the target DNA (usually genomic DNA of interest). After synthesis of the probe, both the probe and target DNA are denatured and incubated to allow annealing of the probe to target DNA. Oligonucleotide probes modified with fluorophores can be visualized directly because of the fluorescence emitted by the probes ( Fig. 136.6 ); alternatively, antigen-modified oligonucleotides can be visualized by immunologic or enzymatic reactions. Structurally normal target DNA will exhibit a signal at the expected site in the chromosome, whereas rearranged or deleted target DNA will exhibit a signal in an aberrant location or exhibit no signal at all, respectively.
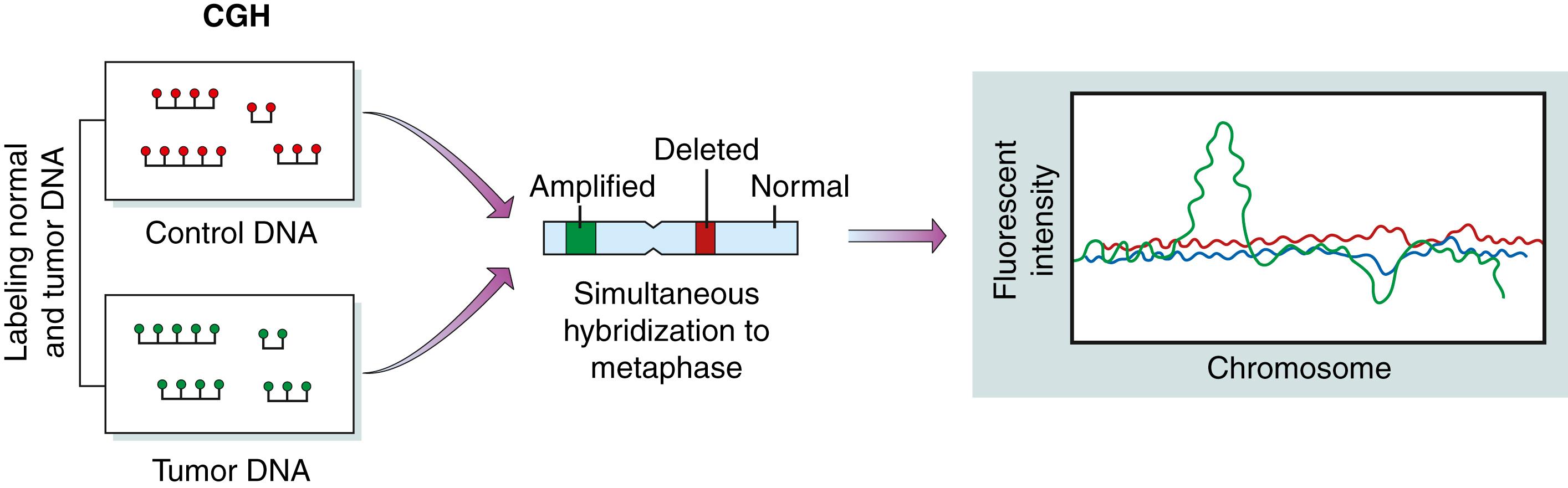
Although FISH has helped identify many chromosomal abnormalities associated with nonsolid tumors, this technique is imperfect for solid tumors. In advanced solid tumors, it is often impossible to interpret tumor chromosome spreads with accuracy because chromosomes are often altered beyond recognition in solid tumors. In comparative genomic hybridization (CGH), which is a variant of FISH, chromosome spreads are generated by differentially labeling test and reference genomic DNA oligonucleotide samples ( Fig. 136.7 ). These can then be applied onto genomic DNA microarrays. CGH arrays provide an unprecedented ability to evaluate chromosomal abnormalities in cancer. In particular, this technique is especially powerful for identification of changes in DNA copy number, such as the increases in gene copy number that occur at the epidermal growth factor receptor (EGFR) locus in a significant number of glioblastomas.
FISH and CGH techniques allow the analysis of gross chromosomal abnormalities (especially changes in copy number or large chromosomal translocations) that occur during cancer development. However, comprehensive analysis of cancer genetics must address other specific events, such as point mutations and the sequential loss of tumor suppressor alleles (loss of heterozygosity [LOH]) because these events are key contributors to the initiation and progression of cancer. Large-scale identification of point mutations and LOH requires the ability to scan large spans of tumor genomes with high resolution.
Single nucleotide polymorphism (SNP) analysis is a powerful molecular genetic tool capable of identifying changes in DNA copy number, point mutations, and LOH. SNPs are nonfunctional point mutations that occur at a frequency of about 1% in the human genome. Because many SNPs have been sequenced, they can be used to compare the haplotypes of cancer genomes with other nontumor DNA from the same individual. Moreover, because the genomic location of SNPs is known, changes in the SNP genotype of tumor DNA identify genomic regions that can be further analyzed by PCR to detect precise DNA aberrations (e.g., point mutations, deletions, insertions) that underlie the changes in SNP haplotype. Microarray platforms have made it possible for up to 500,000 SNPs to be analyzed in a single experiment, allowing excellent genomic resolution.
CGH, FISH, and SNP techniques identify changes in genomic DNA. Although these platforms can be used to make inferences about gene expression, they do not provide direct data about the transcription of these genes from DNA to messenger RNA (mRNA). The importance of studying changes in gene expression is aptly represented by the expression pattern of vascular endothelial growth factor (VEGF) during the genesis of glioma. Although the VEGF gene is rarely mutated in GBM, VEGF mRNA and protein levels are often markedly elevated, which leads to an increase in tumor angiogenesis and virulence.
Gene expression arrays are now widely available. In practice, gene expression array techniques are identical to the CGH and SNP arrays, except that cDNA (transcribed from mRNA) is most frequently used as a substrate for hybridization. Currently, up to 19,000 unique genes can be analyzed for their expression levels in a single array experiment.
Over the past decade, as our understanding of gene expression and its regulatory mechanisms has evolved, equal attention has been given to the study of microRNA molecules and final protein products. MicroRNAs are a class of noncoding RNA molecules that bind to target mRNA and regulate the protein expression posttranscriptionally. Comparative studies of microRNA arrays between tumor cells and normal cells help shed light on differences in gene expression, which would not be evident simply from cDNA microarrays. Similarly, proteomics allows large-scale comparative analysis of proteins, most often using mass spectroscopy. This approach allows direct comparison of protein expression identity and quantity between tumor cells and normal cells.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here