Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The identification of infection in biopsied tissues is the primary responsibility of the surgical pathologist. In an age when both noninvasive and minimally invasive approaches and techniques have increased, it is important to revisit the role of the biopsy in the diagnosis of infection ( Box 2.1 ). Isolating microorganisms in the microbiology laboratory is a sensitive and accurate approach to their identification, but it has several important limitations. First, it cannot distinguish infection from colonization, nor can it ascertain the significance of the isolated organism. Only the presence of an organism in situ, together with an expected inflammatory response by the host, constitutes acceptable evidence of its role in infection.
Establish morphologic diagnosis of infection
Assess immunocompetence of the host
Narrow the differential diagnosis of possible pathogens
Confirm results of microbiologic cultures
Refute the relevance of microbiologic cultures
Establish diagnosis unrelated to infection
Identify concomitant infection in a primary inflammatory or neoplastic disorder
Identify new pathogens
For example, consider how to interpret the clinical significance of a fungus isolated from the airways of a patient with bronchiectasis who also has a new pulmonary infiltrate in the setting of immunosuppression. Is the fungal isolate the likely cause of the opportunistic infection, or might it be a benign commensal? Studies have attempted to address this question with guidelines formulated for practice, but these are indeed merely “guidelines” because only identification of a potential pathogen within a site of infection can provide substantive evidence that the fungus is an invasive pathogen. For this and other reasons to be addressed in this text, the pathologic diagnosis of infection is a critical element in formulating optimal therapy.
Tissue sampling is fundamentally important in the diagnosis of infection. All excised tissues should be considered as potentially infective. This approach fosters due diligence with respect to the possibility of contagion, as well as thoughtful concern as to how the tissues will be handled to optimize the chances of establishing an accurate diagnosis ( Box 2.2 ). Samples of excised tissues should be harvested by sterile technique and sent to the microbiology laboratory with information concerning the types of organism that are being considered diagnostically. Directions to consider anaerobic and fastidious species should be clearly stated.
Make touch imprints for histochemical staining
Handle samples for microbiologic culture with sterile technique
Harvest samples for ultrastructural examination in glutaraldehyde fixative
Harvest fresh samples for appropriate polymerase chain reaction assays
Freeze portion of biopsy specimen for research
After all of this is done, place biopsy specimen in formalin
The surgical pathologist must ascertain that all diagnostic possibilities have been considered. Consultation with an infectious disease specialist can be invaluable in ensuring that specimens are properly handled ab initio. What must be avoided is thoughtlessly placing a biopsy specimen directly into formalin fixative without first considering a diagnosis of infection.
Touch imprints should be routinely prepared and can be stained in the frozen-section suite or in the microbiology laboratory. In general, 5 to 10 touch imprints will suffice, with sampling from the most suspicious portions of the biopsy specimen (e.g., areas of necrosis or suppuration).
Harvesting a portion of the biopsy specimen for ultrastructural analysis can foster the accurate diagnosis of many organisms (e.g., viruses, Tropheryma whippeli, microsporidia). Specimens may be harvested for polymerase chain reaction (PCR) testing to establish the diagnosis of others (e.g., Coxiella , Mycobacterium , Rickettsia ).
The rapid diagnosis of a frozen section can help to focus on the diagnostic workup. All of the pertinent histochemical and ancillary studies can ideally be ordered before the permanent sections are processed, to avoid undue delay in diagnosis.
Because host immune mechanisms can greatly amplify the host response, the actual number of pathogens present in tissues is frequently surprisingly small. This means that many sections may need to be examined before a pathogen is identified. Although few surgical pathologists would balk at the idea of ordering additional sections to exclude malignancy in a biopsy they deemed suspicious, it is not uncommon for a pathologist to examine only a single histochemically stained tissue section in the diagnostic process of infection. More egregious is the fantasy that the causative infectious agent will eventually be diagnosed by the microbiology laboratory, so there is no need for the surgical pathologist to belabor the process.
This approach is wrong minded for several reasons. First, the microbiology laboratory may fail to identify a causative organism. Second, the organism isolated by the laboratory may not represent the actual infective agent in vivo. The analogy is the role for Gram staining of secretions in chronically intubated patients to determine whether there is a neutrophilic exudate consistent with infection and whether there is a predominating organism—steps that can promote the choice of appropriate antibiotic therapy. In this setting, undue emphasis on culture results can lead to a seemingly endless process of adding or eliminating antibiotics in patients who are merely colonized by bacteria and not actually infected. Treatment decisions that do not take into account the host response and dominating organisms will tend to favor the production of increasingly antibiotic-resistant isolates and may potentially compromise public health. This is only one of several compelling reasons to consider diagnostic biopsies in patients with infections in situations that do not readily yield to noninvasive approaches.
Despite the merits of examining biopsy specimens in the diagnosis of infection, one must be aware of those situations in which the sensitivity and specificity of histochemically stained sections are limited. An example is tuberculosis, in which biopsies can fail to demonstrate mycobacteria in almost half of cases. However, even in this setting, the appearance of the inflammatory response in situ should foster a working diagnosis that is often sufficiently reliable to institute empirical treatment.
There is currently no uniformly accepted classification schema for the histologic patterns of response yielded by microorganisms. The inflammatory response in infection is a function of the host response, which is in turn a function of (1) the anatomy of the affected organ, (2) the virulence factors produced by the infective agent, and (3) host immunocompetence. The surgical pathologist must be aware that a single species of microorganism may be capable of evoking a variety of different patterns of inflammation. An example is the broad spectrum of disorders produced in response to infection with Aspergillus spp., which ranges from benign colonization, to hypersensitivity responses, to malignant angioinvasive infection.
The characteristic types of inflammation elicited by infection ( Table 2.1 ) can be broadly categorized as follows.
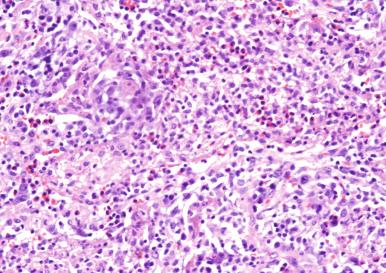
Pyogenic responses. In these responses, neutrophils predominate, leading to pus formation. They are evoked primarily by bacteria, although viruses and fungi can also elicit them ( Fig. 2.1 ).
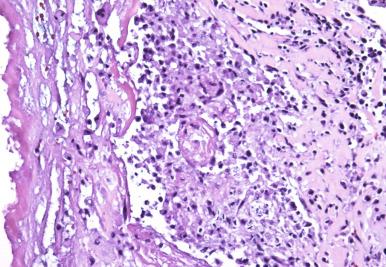
Necrotizing inflammation. Tissue necrosis can occur in several forms. In certain infections, such as those caused by amoebas or gram-negative bacteria, liquefactive necrosis is frequently seen ( Fig. 2.2 ). Other forms, such as ischemic, mummefactive, and caseous necrosis, are often seen in mycobacterial and fungal infections.
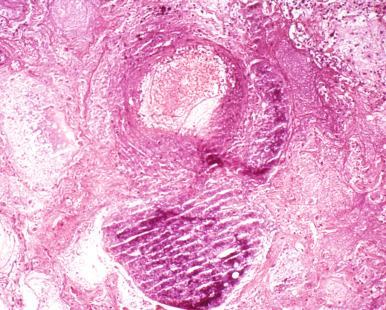
Granulomatous inflammation. This response is characterized by the presence of epithelioid macrophages with multikaryon (giant cell) formation. It appears to reflect cell-mediated immunity to poorly catabolized antigens and is evoked by mycobacteria, fungi, and parasites ( Fig. 2.3 ).
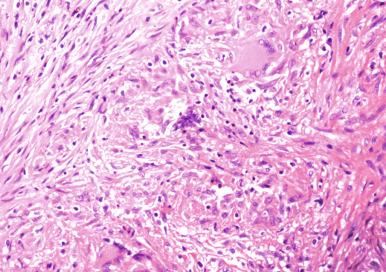
Histiocytic inflammation. These responses are characterized primarily by the presence of foamy macrophages and are a prominent component of infections caused by Legionella, Rhodococcus, Calymmatobacterium, Leishmania, and T. whippeli ( Fig. 2.4 ). In patients who are severely immunocompromised, organisms that normally elicit granulomatous inflammation may instead evoke histiocytic infiltrates.
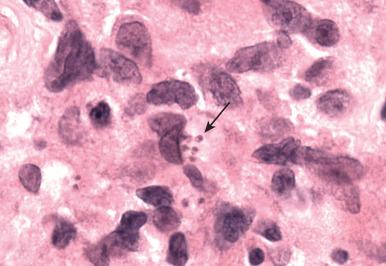
Eosinophilic inflammation. This is seen in response to multicellular parasites and certain fungi ( Fig. 2.5 ).
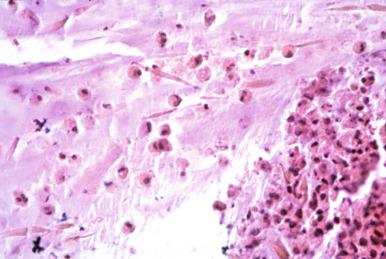
Cytopathic changes. Although this is not properly a type of inflammation, cytopathic changes do reflect a response to viral infection. Nuclear inclusions are part of the response to DNA viruses, whereas cytologic inclusions are seen with some RNA and DNA viral infections, such as cytomegalovirus ( Fig. 2.6 ).
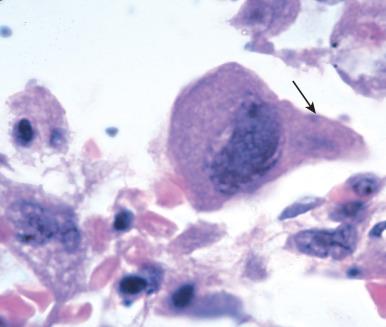
Null responses. In the setting of profound immunosuppression, one may not see inflammation; only the uninhibited growth of microorganisms is apparent ( Fig. 2.7 ).
| Type of Inflammation | Example |
|---|---|
| Exudative inflammation | Pyogenic bacteria |
| Necrotizing inflammation | Gram-negative bacteria, amebiasis |
| Granulomatous inflammation | Mycobacteria, fungi |
| Histiocytic inflammation | Rhodococcus, Legionella, Whipple disease |
| Eosinophilic inflammation | Fungi, parasites |
| Cytopathic changes | Viruses |
| No response | Host energy |
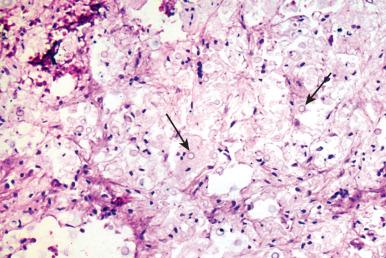
This classification schema is only a crude approximation because overlap patterns of inflammation are common, as with necrotizing granulomatous inflammation, granulohistiocytic inflammation ( Fig. 2.8 ), and granulomatous inflammation with tissue eosinophilia ( Fig. 2.9 ). The primary didactic element is that careful consideration of the histologic response in situ can help to narrow what would otherwise be a very broad differential diagnosis and can also provide invaluable information concerning host immunocompetence. For this reason, surgical pathologists must develop expertise concerning the inflammatory patterns that can accompany reduced immunocompetence resulting from genetic factors, age, toxins, and drugs, because they can skew the expected pattern of inflammation and at times confound the diagnosis.
The identification of microorganisms in biopsy samples is enhanced by the selective application of widely available histochemical stains ( Table 2.2 ). Pathologists should be aware of the spectrum of histochemical staining by microorganisms and knowledgeable with respect to how to formulate combinations of stains to enhance diagnostic specificity.
| Organism | Staining Characteristics |
|---|---|
| Viruses | |
| Influenza | No cytopathic change |
| Coronavirus (SARS) | No cytopathic change |
| Adenovirus | H&E (smudge cells); IHC |
| Cytomegalovirus | H&E (intranuclear and cytoplasmic inclusions); IHC; PAS and GMS (intracytoplasmic inclusions) |
| Herpes virus | H&E (intranuclear inclusions); IHC |
| Measles | H&E (intranuclear inclusions, polykaryons) |
| Respiratory syncytial virus | H&E (polykaryons); IHC |
| Parainfluenza | H&E (intracytoplasmic inclusions) |
| Bacteria | |
| Gram positive | Tissue Gram, GMS (all) |
| Gram negative | Tissue Gram, GMS (some) |
| Legionella | Silver impregnation |
| Nocardia | Tissue Gram, GMS, modified ZN |
| Actinomyces | Tissue Gram, GMS |
| Mycobacteria tuberculosis | ZN and modified ZN; PCR |
| Atypical mycobacteria | Mo dified ZN, ± ZN, PCR |
| Fungi | |
| Histoplasma | GMS, PAS |
| Cryptococcus | H&E, GMS, PAS, mucicarmine; Fontana, IHC |
| Blastomyces | H&E, GMS, PAS, mucicarmine (weak) |
| Coccidiomyces | H&E, GMS, PAS |
| Candida | H&E, GMS, PAS, Gram stain; IHC |
| Aspergillus | H&E, GMS, PAS, IHC |
| Zygomyces | H&E, GMS, PAS |
| Pseudeallescheria | H&E, GMS, PAS |
| Alternaria and dematiaceous fungi | H&E, GMS, PAS, Fontana |
| Parasites | |
| Protozoa | H&E, PAS, Gram stain (microsporidia); IHC (Toxoplasma) |
| Metazoans | H&E, trichrome stain |
| Echinococcus | GMS in chitinous wall, modified ZN (hooklets) |
| Paragonimiasis | Ova birefringent |
| Schistosomiasis | Lateral and terminal spines stain with modified ZN |
The majority of pathogens can be identified with the standard hematoxylin and eosin (H&E) stain. These include cytopathic viruses, some bacteria, most fungi, and virtually all parasites ( Box 2.3 ).
Cytopathic viruses
Bacteria in colonies or in “granules”
Most fungi
Parasites
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here