Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Over the past 2 decades, advances in next-generation sequencing have demonstrated that even though most of the human genome is transcribed into RNAs, only 1% to 2% of the genome encodes for proteins. Therefore RNAs can be grouped into two major classes: (1) coding RNAs and (2) noncoding RNAs (ncRNAs), which account for the majority of RNAs in humans. Although ncRNAs were originally considered “evolutionary junk,” it is now evident that they perform critical regulatory functions on diverse cellular processes, ranging from development to cell growth. , Furthermore, the amount of ncRNAs correlates with the complexity of the organisms because ncRNAs exert significant influence on the organization of higher structured animals.
NcRNAs are divided into two main classes based on their size. Short or small ncRNAs are smaller than 200 nucleotides (nt) whereas long ncRNA (lncRNAs) are longer than 200 nt. The most known class of small ncRNAs are microRNAs (miRNAs), which are around 22 nt.
miRNAs are endogenously expressed ncRNAs of around 22 nt in length. The initial identification of miRNAs was in 1993 when two groups of investigators reported that small transcripts of the lin-4 gene in C. elegans posttranscriptionally regulated the lin-14 gene, suggesting that lin-4 regulates lin-14 translation via an antisense RNA-RNA interaction. , These landmark findings began the era of miRNAs in the biological sciences. Since then, the significance and essential roles of miRNAs in biological processes have been intensely studied. Investigators have shown that miRNAs participate in nearly all developmental and pathologic processes. Importantly, some studies suggest that miRNAs may represent potential biomarkers and therapeutic targets for human diseases. , ,
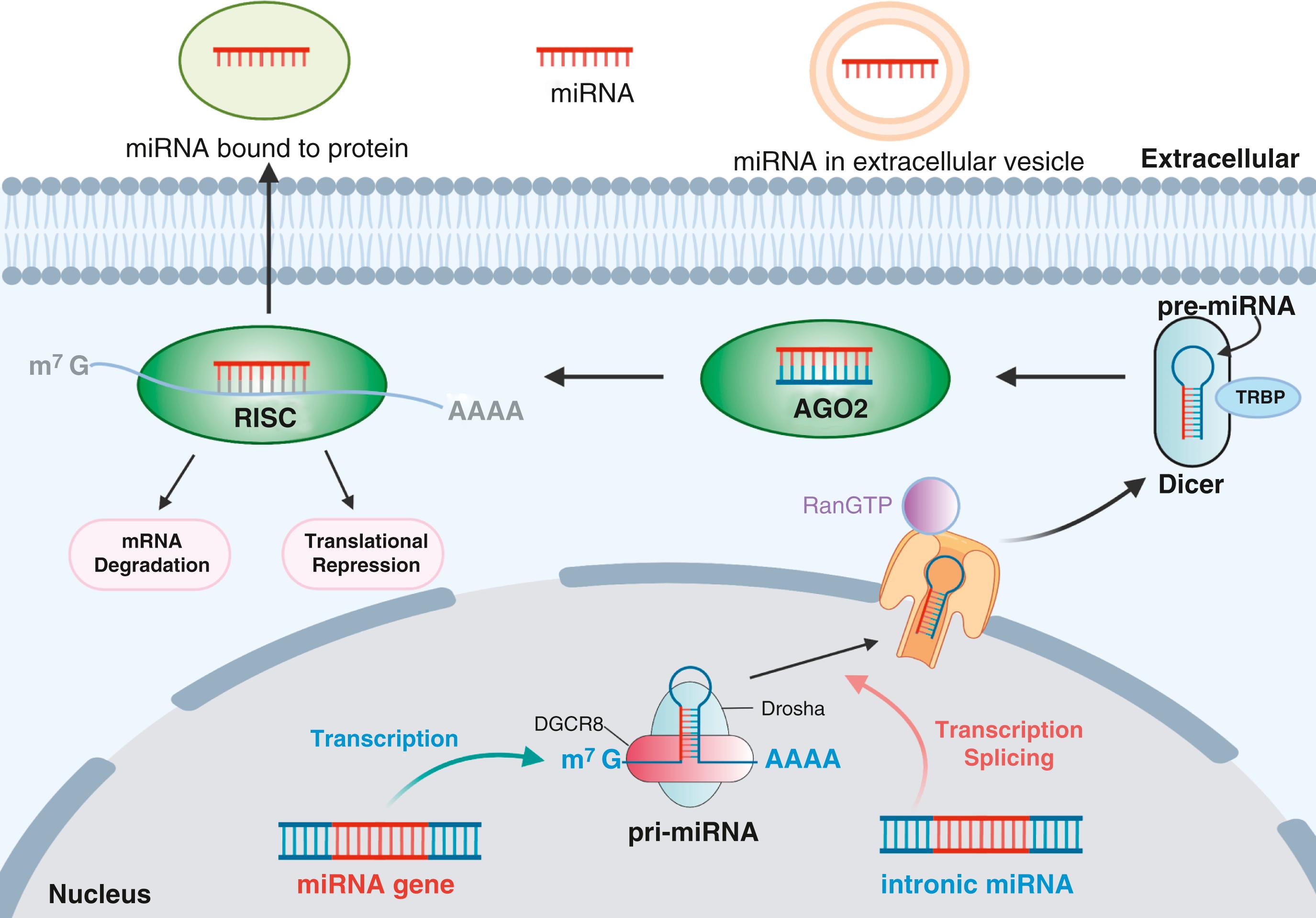
Genes encoding miRNAs constitute one of the most abundant gene families. They are widely distributed in animals, plants, protists, and viruses. , To date, more than 2600 miRNAs have been identified in the human genome according to the most recent release of the miRNA database (miRBase). , miRNAs are encoded by introns of noncoding or coding transcripts, as well as by their own genes. Additionally, they can be transcribed either individually or in clusters as polycistronic transcripts.
After RNA polymerase II–mediated transcription to form primary miRNAs (pri-miRs), usually over 1 kb in length, the maturation of miRNAs occurs through two steps of endonucleolytic cleavage ( Fig. 24.1 ). Pri-miRs are cleaved in the nucleus by the microprocessor, a protein complex primarily composed of the RNase III enzyme ribonuclease 3 (also known as RNASEN or Drosha) and its cofactor, DiGeorge syndrome critical region gene 8 (DGCR8 or Pasha). , The hairpin loop is first recognized by DGCR8, which recruits RNASEN to cut the pri-miR at the base of the stem structure, subsequently releasing the stem-loop portion of around 65 nucleotides termed precursor miRNAs (or pre-miRNAs). The pre-miRNAs then translocate out of the nucleus via the adenosine triphosphate (ATP)-dependent Exportin-5 protein (Xpo5). , In the cytoplasm, pre-miRNAs interact with a second RNase III enzyme, Dicer, and its cofactor, HIV-1 transactivating response (TAR) RNA-binding protein (TRBP). , Dicer cleaves off the loop structure, leaving only the stem region referred to as the miRNA duplex , because the two strands of the stem are no longer covalently connected and are instead only held together by base pairing. This allows the duplex to unwind and separate in an ATP-independent fashion by Dicer, TRBP, and argonaute 2 (AGO2), which collectively form the RNA-induced silencing complex (RISC). , Through the interaction of the miRNA duplex and RISC, the duplex unwinds, which allows one strand, the guide strand, to load onto AGO2. The other strand, the passenger strand, is degraded. The guide strand is the mature and fully functional miRNA unit and can target a variety of coding messenger RNAs (mRNAs) to promote degradation and translational repression.
A hallmark of miRNAs is their tremendous potential to target a large number of genes, often in common pathways. With an estimated 60% of human mRNA having conserved miRNA targets, miRNAs represent one of the key regulators and informative biomarkers of a set of genes controlling different cellular processes rather than single genes. , To comprehensively evaluate the influence of miRNAs on multiple proteins by the integration of various downstream genes, the global evaluation of miRNA in the signaling cascade becomes essential.
Long ncRNAs (lncRNAs) are found in diverse species ranging from viruses, prokaryotes, and plants to humans and can regulate gene expression at multiple levels in the cell at both transcriptional and posttranscriptional levels through their interactions with nucleic acids and proteins in both a sequence-specific and structure-specific manner. , Moreover, lncRNAs are exquisitely regulated under different physiologic conditions and in different human diseases. Nevertheless, our current understanding of lncRNAs is lacking compared with miRNAs. This is because of the fact that unlike miRNAs, lncRNAs are poorly conserved among different species, which therefore limits the use of animal models for the investigations of lncRNAs.
miRNAs in the heart have been extensively studied in the past decade. Cardiac-specific miRNAs play critical roles in heart development, myocardial regeneration, and cardiac remodeling. miR-1 and miR-133 are muscle-specific miRNAs representing the most abundant miRNAs expressed in the heart. , Mouse embryos lacking Dicer in the developing heart die of cardiac failure before birth and exhibit poorly developed ventricular myocardium. Dicer is necessary for cardiac outflow tract alignment and chamber septation. Conditional Dicer gene deletion in 3-week-old mice provokes premature death and induces rapid and dramatic biventricular enlargement accompanied by hypertrophy, myofiber disarray, ventricular fibrosis, and strong induction of fetal gene transcripts in adult mice. Cardiomyocyte-specific deletion of DGCR8, an essential partner of Drosha, in newborn hearts induces dilated cardiomyopathy and heart failure (HF). Several miRNAs, including miR-1, miR-133, miR-208a, miR-208b, miR-499, the miR-15 family, and the cluster of miR-17 to miR-92 miRNAs, have been reported to be critical for cardiac development. ,
miRNAs are also essential in myocardial regeneration. The global loss of miRNAs in embryonic stem cells (ESCs) by deletion of Dicer and Drosha or DGCR8 leads to defective differentiation and proliferation. , Muscle-specific miR-1 and miR-133 have been identified to influence muscle lineage commitment and regulate muscle proliferation and differentiation. , One study analyzing the roles of miR-1 and miR-133a in the differentiation of three types of pluripotent stem cells shows the importance of miR-1 and miR-133a in controlling the cardiac fate of the pluripotent cells. miR-499 is a cardiac-specific miRNA and is essential for cardiac commitment of human ESCs. miRNAs are also potent regulators of cell fate reprogramming and several miRNAs, including miR-1, mi-133a, mi-208a, and mi -499, have been shown to be capable of converting cardiac fibroblasts into cardiomyocytes. ,
Studies have shown strong evidence for the critical roles of miRNAs in the regulation of cardiac ion channels and transporters. miR-1 - 2 deletion in mice induces abnormal cardiac electrical activity, including bradycardia, shortened PR interval and prolonged QRS complexes with increased incidence of sudden death. Abnormality in cardiac excitability may partially result from the upregulation of Iroquois Homeobox domain 5 (Irx5), which is a confirmed target of miR-1. Irx5 is a known homeodomain transcriptional inhibitor of the Kcnd2 gene encoding for the pore forming K V 4.2 subunit of the transient outward K + channel. As a result, the expression of Kcnd2 is downregulated, leading to decreased transient outward K + currents (I to ).
miR-1 has been shown to target Kir2.1 channel, and overexpression of miR-1 suppresses the expression of Kir2.1 channel and connexin43. miR-1 also inhibits the expression of other ion channel genes, including Scn5a, Cacna1c, Kcna5, and Kcne1. PP2A regulatory subunit, B56alpha, is a target of miR-1, and miR-1 overexpression increases phosphorylation of L-type Ca 2+ channels and ryanodine receptor 2 (RyR2) by suppressing PP2A, which enhances RyR2 activity and cardiac excitation-contraction coupling.
In contrast, miR-133 has been reported to depress PP2A activities, resulting in abnormal RyR2 function and Ca 2+ cycling in HF, leading to an increased propensity for cardiac arrhythmias. In addition, miR-133 may also regulate hyperpolarization-activated cyclic nucleotide-gated channels (HCN2 and HCN4). miR-208 is a cardiac-specific miRNA and an analysis of mice lacking miR-208a indicates that miR-208a is required for proper cardiac conduction and expression of the gap junction protein connexin40. miR-135a has been reported to regulate the expression of the Na + /Ca 2+ exchanger and affects the Ca 2+ signaling and automaticity. The upregulation of miR-30d in atrial fibrillation (AF) patients resulted in the suppressed expression of a G protein–gated K + channel (I KACh ) in atria. One study has shown that miR-221 and miR-222 target Cacna1c and Kcnj5 (G protein–activated inward rectifier K + channel), leading to an altered expression of the L-type Ca 2+ channel, Kcnj5 channel, and their current densities.
One recent study examines the roles of Dicer1, a critical enzyme processor, for the maturation of miRNAs in the heart. Delivery of a Cre recombinase expressing adenovirus (Ad-GFP-Cre) to the left ventricles of a conditional Dicer1 knockout mouse model, ( Dicer1 tm1Bdh /J ), which contains a floxed RNase III domain critical for Dicer1, results in cardiac-specific knockout of the Dicer1 enzyme. Similar to miR-1 - 2 deletion, knockout of Dicer1 in adult mouse cardiomyocytes results in decreased I to and prolonged action potential duration (APD). In addition, an increased Ca 2+ current (I Ca ) is observed. This model allows the evaluation of the roles of miRNAs on ionic currents without the interference of organ level changes, such as hypertrophy or fibrosis. Parallel experiments in neonatal Dicer1 tm1Bdh /J mouse cardiomyocytes by transduction of Ad-GFP-Cre demonstrate increased transcripts for the Ca V 1.2 (α 1c ) pore-forming subunit of the Ca 2+ channel ( Cacna1c ) and decreased transcripts for the K V 4.2 ( Kcnd2 ). Moreover, there is a significant increase in the expression of the known Kcnd2 transcriptional inhibitor, Irx5, a validated target of miR-1. , , Taken together, these studies support the critical role of miRNAs in the regulation of cardiac ion channel expression.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here