Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
High-resolution genome analysis techniques are now being used in international cancer genome analysis efforts to catalog aberrations driving the pathophysiology of nearly all major cancer types. The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/ ) project and the International Cancer Genome Consortium (ICGC, http://www.icgc.org/ ) represent the largest of these. The TCGA project is assessing aberrations in 500 to 1000 tumors from each of about 20 major human cancer types and together the TCGA and ICGC are currently committed to the analysis of ∼47 separate tumor lineages. Results for glioblastoma, serous ovarian cancer, colon cancer, squamous cell lung cancer, and breast cancer are already available from the TCGA project, and other technology centers around the world have contributed studies of cancers of the breast, pancreas, prostate, lung, and kidney and of melanoma, myeloma, and AML. The overall intent of these international genomics efforts is to provide a knowledge base of cancer genome landscapes that can be used to develop more effective cancer management strategies—for example, by enabling early detection of lethal or recurrent cancers, diagnosing patients with low malignancy potential tumors who may be spared from aggressive treatment, identifying novel therapeutic targets, and assigning therapies to the patients in which they are most likely to be effective. We summarize in this chapter general and specific aspects of human cancers that are emerging from international genomics efforts, describe computational and experimental efforts to identify aberrations that contribute to aspects of human cancer pathophysiology, and illustrate how genomic information is being used in aspects of cancer management.
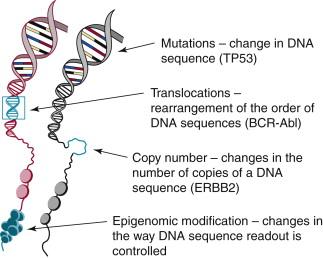
Genome aberrations found to be important in human cancers are illustrated in Figure 24-1 and include (1) somatic changes in copy number that increase or decrease the levels of important coding and noncoding RNA transcripts, (2) somatic mutations that alter gene expression, protein structure, or protein stability and/or change the way transcripts are spliced, (3) structural changes that affect transcript levels by altering gene-promoter associations or create new fusion genes, and (4) epigenomic events that alter transcription levels of diverse signaling pathways and that enable rapid emergence of therapeutically resistant subpopulations. Table 24-1 shows the number of gene aberrations catalogued in the Cancer Gene Census maintained by the Sanger Institute that have been implicated in one or more human cancer types (see http://www.sanger.ac.uk/genetics/CGP/Census ). These aberrations have been implicated in deregulation of pathways that influence all aspects of cancer progression including invasion, immortalization, DNA replication and repair, proliferation, apoptosis, angiogenesis, motility, and adhesion. However, the 487 genes implicated in the Cancer Gene Census are likely to be only a small fraction of the aberrant genetic events that play important roles in the form and function of human cancers. These genes have been selected primarily because they were implicated in model system studies or because they occur frequently in one or more cancer subtypes. However, recent genomic studies demonstrate that hundreds to thousands of genes may be affected by somatic mutations and epigenomic modifications in an individual tumor. These comprise a mix of “driver” aberrations and a usually larger number of “passenger” aberrations. Driver aberrations are genomic or epigenomic events that are selected during tumor progression because they alter one or more aspects of cell and tissue physiology to allow cancer initiation, progression, and/or dissemination. Table 24-2 , for example, illustrates several genes that might contribute to the deregulation of the cancer hallmarks designated by Weinberg and Hanahan. Some of these aberrations have been targeted by therapeutics, and others have been shown to influence clinical behavior, including response to targeted and nontargeted therapies. Passenger aberrations do not contribute to cancer pathophysiology but arise by chance during progression in a genomically unstable tumor and are carried along because they exist in cells carrying driver aberrations. Driver aberrations are identified either because they occur frequently in tumor subpopulations or because they have been identified as contributing to aspects of cancer pathophysiology in laboratory models of cancer—for example, cultured cancer cells or genetically manipulated nonmalignant cells, or cancer cells grown as xenografts in animals—or have been demonstrated to influence aspects of cancer in genetically engineered living organisms. Several general cancer genomic observations from international cancer genomics projects are summarized in the following paragraphs.
| Aberration Type | Number of Aberrations | Examples of Prominent Affected Genes |
|---|---|---|
| Amplification | 16 | ERBB2, EGFR, MYCN, MDM2, CCND1 |
| Frameshift mutation | 100 | APC, RB1, ATM, MLH1, NF1 |
| Germline mutation | 76 | BRCA1/2, TP53, ERCC2, RB1, VHL |
| Missense mutation | 141 | ARID1A, ATM, PIK3CA, IDH1, KRAS |
| Nonsense mutation | 92 | CDKN2A, FANCA, PTCH, PTEN |
| Other mutation | 26 | BRAF, PDGFRA, PIK3R1, SOCS1 |
| Splicing mutation | 63 | GATA3, MEN1, MSH2, TSC1 |
| Translocation | 326 | ABL1, ALK, BCL2, TMPRSS2, MYC |
| Cancer Hallmark | Aberrant Gene |
|---|---|
| Resisting cell death | BCL2, BAX, FAS |
| Genome instability and mutation | TP53, BRCA1/2, MLH1 |
| Inducing angiogenesis | CCK2R |
| Activating invasion and metastasis | ADAMTSL4, ADAMTS3 |
| Tumor-promoting inflammation | IL32 |
| Enabling replicative immortality | TERT |
| Avoiding immune destruction | HLA loci, TAP1/2, B2M |
| Evading growth suppressors | RB1, CCND1, CDKN2A |
| Sustaining proliferative signaling | KRAS, ERBB2, MYC |
| Deregulating cellular energetics | PIK3CA, PTEN |
One important observation from many genomic studies is the existence of recurrent molecular features that allow cancers that occur in specific anatomic regions to be organized into subtypes. The subtypes likely arise in distinct cell types within each tissue and are different diseases that differ in clinical outcome and/or response to therapy. Early genomic studies relied on expression patterns for cancer subtype definition, but current strategies use multiple data types (e.g., genome copy number, mutation, and expression) for subtype definition. Interestingly, epithelial and mesenchymal subtypes appear to be present in tumors that are of epithelial origin. The mesenchymal-like cancers tend to be more rapidly proliferating and motile and associated with reduced survival duration. Some tumor types show remarkably high transcriptional similarity, for example, in triple-negative breast cancer and high-grade serous ovarian cancers. Many genomic aberrations also appear in multiple tumor subtypes. Some of the most common aberrations observed in multiple tumor types include amplifications of MYC and EGFR, deletion of CDKN2A and PTEN, and mutation of TP53 and PIK3CA. For a more comprehensive assessment, Kim and colleagues summarize recurrent genome copy number aberrations in 8000 cancers. Efforts are now under way to combine data types (e.g., expression, genome copy number, and mutations) to increase the number of subtypes in order to increase the precision with which patients can be stratified according to outcome and/or therapeutic response. Of course, this divides cancers into increasingly smaller subpopulations, so very large numbers of samples are needed to establish subtype differences in treatment response or overall outcome.
The number of aberrations that are present in an individual tumor can be remarkably high. The somatic mutation rate in human cancers varies between cancer types from about 0.1 to 10 mutations per megabase, but individual tumors may carry as few as a hundred to more than a million somatic aberrations. High genomic instability occurs because of loss of telomere function during progression in the absence of telomerase, diminished DNA repair capacity resulting from genomic and epigenomic deregulation of DNA repair pathways, increased damage resulting from oncogene-induced oxidative stress, and toxic environmental exposures. In some cases, the exact DNA sequence change in a mutation reflects the type of agent that causes the cancer—for example, mutations in sun-related cancers show CC to TT mutations caused by UV-induced cytosine dimers, whereas smoking-induced cancers in the lung are characterized by G→T transversions caused by the polycyclic aromatic hydrocarbons in tobacco smoke. Ultimately, the functions and/or expression levels of hundreds to thousands of genes may be altered in an individual tumor. An unknown number of these will be drivers. Among these, some will have a strong, possibly dominant influence on an individual tumor, whereas others may have a more modest or near-negligible impact. So far, most attention in the field has focused on the strong drivers. However, it seems likely that the ensemble of aberrations will have to be taken into account in explaining the overall behavior of an individual tumor, which is addressed in a later section.
The same drivers of genome instability that enable tumor development also operate during tumor progression. As a result, individual tumors become increasingly heterogeneous as distinct clonal populations within the tumor evolve in diverse microenvironments, producing highly branched lineages. For example, events that enable metastasis may occur late during the genetic evolution, whereas mutation of TP53, a key player in genome stability, can be an early event. These instabilities and the resultant intratumor heterogeneity in an individual tumor are likely responsible for the rapid evolution of therapeutic resistance. This heterogeneity complicates clinical decision making because the importance of a low-frequency but actionable aberration remains unclear. One possible way forward is to focus treatment on aberrations that occur early during tumor development. The order in which aberrations occur can be inferred by examining a tissue at various stages of disease progression by serial sampling of clinical tissue from individual patients, by computational methods that examine mutation frequency, or in some cases by analysis of the interactions between mutations and copy-number abnormalities.
Transforming cancer genomic data into interpretable knowledge consists of finding the parts and learning how they work together to enable aspects of cancer pathophysiology. Hypothesis-driven research has gone quite far in this process, but full understanding will require systematic analysis, both computational and experimental, of the aberrations that occur within a tumor genome.
Computational strategies to identify candidate driver aberrations begin with the cataloging of all aberrations and then move to the selection of high-priority candidate drivers.
Identification of genes that enable aspects of cancer pathophysiology (driver genes) is complicated by the high genomic heterogeneity within and between tumors. Nearly all cancer genomes analyzed to date appear to have at least one driving oncogenic point mutation, and the vast majority show copy number changes over both large chromosomal segments and smaller, more targeted regions of the genome. The evidence for structural rearrangements being a primary cause in most tumor types is less clear, but diseases including many leukemias, lymphomas, sarcomas, and prostate cancers all incontrovertibly show that rearrangements can be critical ( http://atlasgeneticsoncology.org ). Changes to chromatin state also are partly responsible for many cancers. Over the past 20 years a number of technologies (predominantly microarray based) have been successfully used to catalog cancer genome aberrations, but nearly all efforts now depend on nucleic acid sequencing technology (Mardis, chapter on “The Technology of Analyzing Nucleic Acids”).
Point mutations are identified by aligning DNA sequences obtained from cancer samples to normal genomes using tools such as BWA. The requirement for the normal genome sequence is paramount because of private single-nucleotide polymorphisms (SNPs) that occur about once every 100,000 base pairs, a rate that is about 10 times higher than the mutation rate in most epithelial tumors and 100 times higher than the rate of mutations in childhood cancers such as neuroblastoma. Read depth and read quality are critical factors in determining how well mutations can be called within each patient’s cancer genome. Read quality is the error rate per thousand base pairs of sequence. High quality is usually defined as having fewer than 1 error per 1000 bases of sequence. Read depth (the number of times a position in the genome has been sequenced) for high-quality bases then governs both the false-positive rate caused by sequencing errors and misidentifying private variants as mutations and false negatives caused by not generating sufficient data to observe mutations reliably. The greater the depth, the more confident mutation calls will be. Typically, 30× coverage of the normal genome and 40× to 80× coverage of the tumor produces high-quality results. Increasing read depth is needed for analysis of samples in which the tumor fraction is low because the presence of normal DNA reads dilutes the aberrant reads. Mutation detection is further complicated by intratumor heterogeneity that causes some aberrations to be present in only a small fraction of the tumor cells. Many groups find value in exome sequencing—that is, targeting the small fraction of the genome that is coding, at even deeper levels (for example, 150×). Verifying the sensitivity of mutation calling remains difficult because there are no good true mutation standards.
Detection of insertions and deletions (indels) remains challenging. In principle, the same sequence coverage necessary to find point mutations can be used to identify indels. Unfortunately, the algorithmic methods for indel identification are much more computationally intense. No good estimates exist on how well indel detection software works because of the lack of gold standards against which to measure algorithm performance. In general, indel detection is even more difficult than evaluating the substitution mutations.
Copy number and structural aberrations are identified using a combination of microarray and sequencing approaches. Microarrays and whole-genome shotgun sequencing are capable of identifying changes in DNA copy number that are as small as 1000 base pairs in length. This resolution is sufficiently good that nearly all gene-level aberrations can be detected. Microarray approaches look for differential signal gains from the hybridization, whereas DNA sequences detect changes in read depth. Direct sequencing of genomic DNA represents the most direct way to identify the breakpoints for structural rearrangements, but the methodology is challenging, requiring a high-coverage, high-quality DNA sequence. Often, structural rearrangements cannot be detected with the standard technologies because the sequencing approaches used cannot span the length of repetitive sequences in the human genome. Once a whole-genome shotgun sequence is generated, methods such as BreakDancer and Delly can be used to find the chromosome junctions. Other structural aberration detection technologies are emerging, so it is likely that we will be able to identify the majority of structural breakpoints in the near future.
Detection of promoter methylation is usually accomplished using microarray technologies. Microarrays that can measure methylation at more than 485,000 sites are now commonly used by groups such as TCGA. In principle, DNA sequencing can be used for this purpose, but this is currently economically impractical, with costs 10 to 50 times greater than for microarray approaches. In addition, sequencing approaches currently require unreasonably large quantities of tumor DNA.
RNAseq is now the standard for measuring gene expression. RNA is depleted of ribosomal RNA (rRNA) by either polyA+ selection or any number of rRNA depletion steps and fragmented before complementary DNA (cDNA) production. Short cDNA fragments are sequenced and mapped to the human genome reference. Algorithms to estimate which transcripts are being produced and their relative abundances are used to interpret the fragment data. One strength of RNAseq analysis is that it does not require that the transcriptome be known, and thus it has enabled the study of noncoding RNAs, including lincRNAs and, with adapted protocols, miRNAs. RNAseq methods are still being refined, with improvements in molecular and algorithmic approaches regularly being developed.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here