Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Current DNA sequencing methods differ dramatically from a mere 7 years ago, when next-generation sequencing (NGS) instrumentation was first introduced. Indeed, the science of DNA sequencing is only 35 years old, and its companion discipline known as genomics has been revolutionized by the advent of NGS instrumentation and its application to myriad biological questions. Whereas conventional DNA cloning and sequencing approaches, largely based on the initial descriptions by Fred Sanger and colleagues, provided key reference genome sequences for model organisms (Caenorhabditis elegans , Escherichia coli , Arabidopsis thaliana , Saccharomyces cerevisiae , Mus musculus) and for humans, next-generation instruments and associated analytical efforts have truly revolutionized the nature of biological inquiry. This fact is nowhere more evident than in the study of cancer, first proposed by Bovery as a disease whose origins lay in profound alterations of the nuclear DNA, remarkably before DNA was found to be the hereditary material and instruction set for the organism. His hypothesis about cancer as a disease of the genome was first supported by microscopic observations of reoccurring chromosomal translocations in leukemic cells. Later, the fusion proteins resulting from these translocations were proven necessary and sufficient to induce the developmental arrest and proliferation of leukemic cells. Hence, genomic alterations were a precursor to cancer’s development.
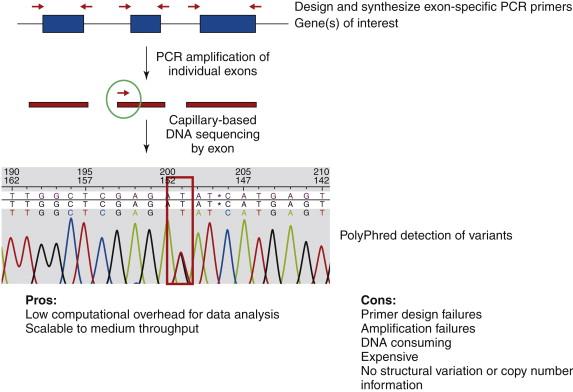
Having the reference human genome in hand, the combination of polymerase chain reaction (PCR) and Sanger sequencing by fluorescent capillary methods provided the first nucleotide-level evidence that point mutations in tyrosine kinase genes also were associated with carcinogenesis. Thus, DNA sequencing was shown to provide a higher resolution “microscope” with which to catalog specific genes that were commonly mutated in cancerous cell genomes. These efforts also established the rationale that genes found recurrently mutated in different cancer samples and ultimately across different types of tumors were likely drivers of oncogenesis. Although increasing the scale of this approach was attainable, there were known limitations to PCR- and Sanger-based approaches ( Figure 23-1 ). On a practical level, PCR had many sources of failure, including the inability to design primers that bind with fidelity in repetitive sequences or exclude pseudogenes or distinguish specific genes in gene families, as well as amplification failure when primer annealing failed due either to DNA polymorphisms or to structural alterations that changed or removed one or both binding sites or to poor amplicon yield from high G+C content (common in the first exons of genes). Furthermore, data generation for large numbers of genes required a significant amount of DNA to be obtained from each tumor. In addition, although large-scale automation could be employed to provide throughput and reproducibility, scaling successfully to address all human genes in large numbers of samples was cost prohibitive. Beyond practical considerations, the approach provided no information about DNA rearrangements or copy number alterations and overall no appreciation of reoccurring events outside the genes.
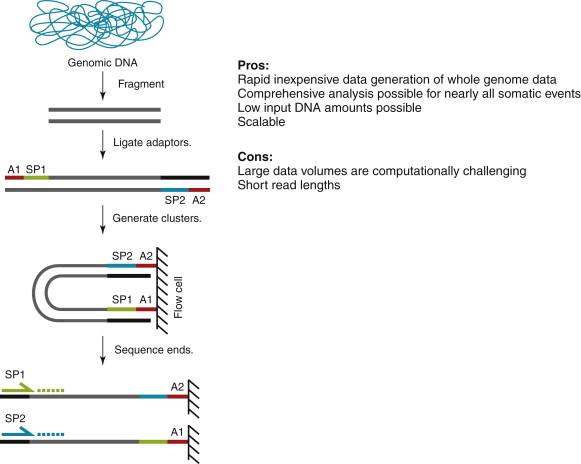
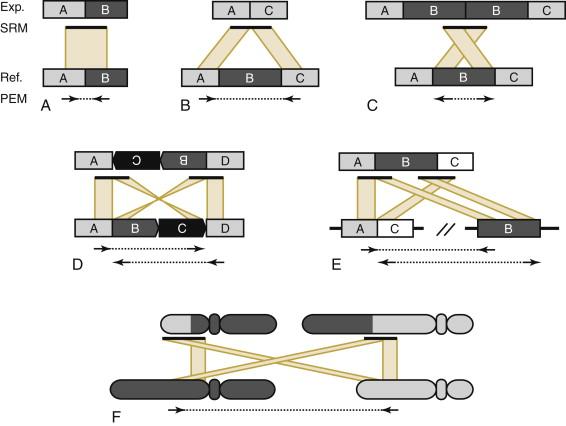
The advent of NGS instruments addressed many of these limitations by providing several advantages in terms of throughput and cost. Although the technical nuances of the various NGS instruments differ, they generally share the same principles ( Figure 23-2 ). These include (1) a simplicity of library construction that requires comparatively little DNA versus PCR-based methods, (2) an enzymatic amplification of each library fragment to produce sufficient signal during the sequencing reaction, and (3) a stepwise sequencing reaction that detects the signal from each nucleotide incorporation reaction of each amplified fragment population before moving to the next reaction. This en masse sequencing approach is why NGS is often referred to as “massively parallel.” Indeed, dramatic increases in scale and speed, and decreases in cost, of data generation by NGS have occurred in a very short time. The size of NGS datasets, especially from whole-genome sequencing (discussed later), coupled with relatively short read lengths compared to Sanger sequencing, have required substantial investments in the development of computer algorithms, computing/IT infrastructure, and computational biology expertise to successfully interpret the data. These computational efforts have been further taxed by continued improvements in NGS data quality (error rates), increasing read lengths, and the ability to generate sequencing data from both ends of each library fragment (typically referred to as “paired end” sequencing) because of the need for new algorithm and data analysis pipeline development as well as refinements to existing computational pipelines. The bottom line is that the first step in interpreting short read data is computational alignment to a reference genome such as the Human Reference Genome. Alignment maps each read or read pair to its origin in the genome and then assigns a quality score to the mapping position that can be interpreted for its certainty of correct placement. Subsequent analytical approaches further interpret the total read alignment (also called “coverage”) in a variety of ways to identify single-nucleotide variants (SNVs), small insertion or deletion events that involve one or a few bases (“indels”), and structural variants (large insertion or deletion events, chromosomal inversions, or translocations–see Figure 23-3 ). Although read length and data quality improvements have overall expanded the utility of NGS for biological experimentation and decreased the cost of data generation by a trajectory that exceeds Moore’s law, the computational requirements of NGS analysis have remained largely unchanged. Many unique aspects, discussed later, further complicate sequencing and analysis from cancer samples. In spite of the obstacles, our understanding of the genomic landscape of cancer has changed remarkably in just a few short years.
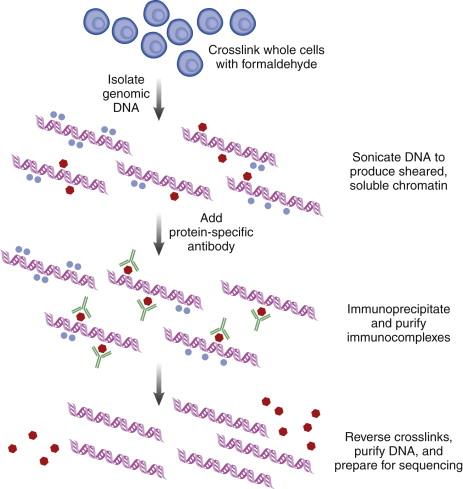
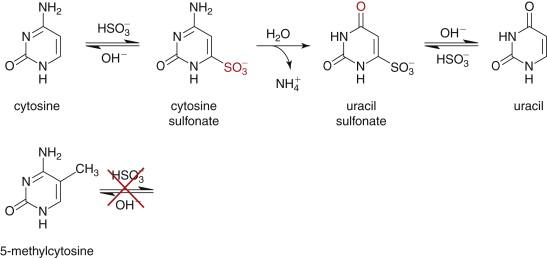
The search for somatic variation in cancer DNA and RNA has a distinct advantage over other complex diseases: The exact comparison of tumor to normal nucleic acids within an individual patient distinctly identifies those alterations that are tumor unique. Furthermore, there are increasing amounts of data from various projects that have begun using NGS methods to catalog large numbers of cancer cases across different tumor types (ICGC [ icgc.org ], TCGA [ cancergenome.nih.gov ], PCGP [ www.pediatriccancergenomeproject.org ] that can be used to inform individual analyses about previously described alterations. In spite of these decided advantages, there are several significant challenges that confound experimental design and analytical approaches in cancer genomics studies. Several examples of these challenges are described next, along with the ways researchers attempt to overcome them, where applicable.
Cancerous cells in solid tumors do not exist in isolation in the body. Rather, they are always in close proximity to normal cells of various types, including stromal cells, immune cells, and components known as the extracellular matrix (ECM). The proportion of tumor cells can be estimated by an experienced pathologist examining the tumor section under hematoxylin and eosin staining, and this estimate is expressed as a “percent tumor nuclei” or “percent tumor cellularity” value. As a result of the association of tumor and normal cells, an isolate of DNA or RNA derived from a solid cancer sample will contain both tumor and normal cells unless a specific procedure such as flow cytometry or laser capture microdissection (LCM) is used first to significantly enrich the percentage of tumor cells in the isolate. Also, certain tumor types, such as those from prostate or pancreas, are more prone to low tumor cellularity. Based on the pathology estimate, decisions in sequencing must be made in the context of tumor cellularity percentages. Namely, if the tumor cellularity is below 60%, the decision must be made either to enrich the tumor by flow cytometry (more common for blood cancers such as lymphoma or leukemia) or by LCM (used for solid tumors), or to try oversampling the tumor NGS library (increased sequencing coverage) by an amount commensurate with the tumor cellularity estimate. Although sorting or LCM seems the most obvious choice, one limitation of either approach is that significantly reduced yields of DNA or RNA will be obtained. Unless specialized procedures are in hand, the low yield may limit the ability to derive high-quality data from such samples. By contrast, oversampling may be effective for DNA sequencing but will be more expensive to generate and will require adjustment of variant calling parameters, or use of a more sensitive variant caller, to effectively identify somatic variants. Oversampling for RNA-seq from a sample with low tumor cellularity is generally not advised, as the tumor transcripts will be too difficult to discern from those of the normal cells unless LCM or sorting is first used to separate the tumor cells from the adjacent normal/nonmalignant cells.
Heterogeneity is a fundamental aspect of cancer cells found within the same tumor of which there are two types, regional and genotypic. Regional heterogeneity reflects the differences that emerge in solid tumors as they grow and progress. It refers to the different regions present in a tumor mass, such as areas of necrosis or areas of invasion (of surrounding normal tissue). Genotypic heterogeneity reflects the fact that cancer cells evolve during the process of tumor progression, so that not all tumor cells share the same somatic genotype. In genotypic heterogeneity, the use of NGS has demonstrated that by comparing the genomes from progression samples (a de novo leukemia compared to its relapse) using high-depth sequencing of somatic mutations, an initiating or “founder” clone can be identified that contains the core mutational load that initiates tumor growth as well as more advanced clones that combine newer mutations with those in the founder clone. One shared aspect of regional and genotypic heterogeneity is that as a tumor mass increases in size, both are more likely to occur in that areas of regional heterogeneity are likely to have genotypic heterogeneity. There are so far only two studies to examine this at the DNA level; one study of two advanced-stage renal cell carcinomas that exhibited extreme genotypic heterogeneity and one study of five early-stage (2/3) breast cancers that showed little to no genotypic heterogeneity when sampled and studied at multiple sites.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here