Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
This chapter includes an accompanying lecture presentation that has been prepared by the author: . ![]()
The basis of CNS malignancies is DNA mutations in genes that regulate cell fate, survival, and genome maintenance.
Most commonly these mutations arise by errors in DNA replication, but initiating mutations can be inherited as a mendelian cancer syndrome.
Approximately two to seven mutations are required for malignant transformation of an individual cancer.
Gene mutations in the most common primary brain malignancy, glioblastoma, are most frequently in genes that regulate telomere maintenance, cell cycle control, and growth factor signaling.
What causes brain cancers? The simple answer is an unfortunate accumulation of DNA damage events, leading to gene mutations and dysregulation of critical normal cell functions. A normal cell will carefully control its growth beyond development or repair/regeneration, but a cancer cell has uncontrolled growth prompted by pathologic instructions from the mutated genome. A normal genome will instruct a cell to coexist with its neighboring cells, and a brain cancer cell with mutations will invade and eventually destroy the surrounding brain.
Alterations to a brain cell genome at critical regions, within genes that control cell growth, cell cycle, and cell death, are the basis for brain tumor formation. How do these mutations arise? In most tumors, the mutations probably arise spontaneously during cell division and/or with failure to properly correct DNA damage. For a small percentage of brain tumors the first mutation is inherited from one of the parents, increasing the risk for brain tumor formation, although the subsequent mutations needed to complete tumor formation are acquired in the same way as in spontaneous tumors.
There are external causes that significantly increase the risk of developing a cancer, but for brain tumors the evidence is limited. High doses of ionizing radiation to the brain have the strongest and best known evidence for increasing the risk of certain brain tumors, as seen with radiation therapy involving the brain, or increases in meningiomas among atomic bomb survivors. , Environmental causes for brain tumor are not as evident. The main carcinogen in our society, tobacco, does not have evidence of producing brain tumor risk as with many other cancers. There are man-made chemicals (e.g., petroleum products, vinyl chloride, fungicides, pesticides) that have been associated with brain cancer in epidemiologic studies, but evidence is often conflicting and more research is needed to demonstrate a direct cause-and-effect relationship. The risk for a primary CNS lymphoma is elevated with immune suppression, and diseases that cause chronic immune suppression indirectly raise the risk of brain cancer. Overall, it can be said that the vast majority of mutations that give rise to brain cancers are spontaneous, and except for hereditary risk of family members for certain rare cancer syndromes or the aforementioned exceptions, it is not possible to predict accurately who in our society is at greater risk for a brain tumor.
There are more than 120 different pathologic classes of brain tumors. For many of the more common of these brain cancers, the underling acquired mutations that drive cancer formation have been identified. Low-cost high-throughput DNA sequencing is commonplace and has been applied to most types of brain cancers. The genes mutated in the development and progression of brain cancer are a restricted set of genes that regulate the fundamental cellular processes of cell fate, cell survival, and genome maintenance. Overall in all cancers there are about 140 of these mutated driver genes identified that cluster within a dozen signaling pathways, and many of these driver mutations have been observed in at least one type of brain cancer.
In this chapter, these commonly altered cancer driver genes are reviewed for the more common brain cancers. Understanding the function and mechanisms of these cancer pathways provides an understanding of how tumors proliferate and provides researchers with better means for targeting tumors, improved diagnosis, and understanding mechanisms of resistance to therapy.
A fundamental concept in cancer is the clonal expansion of tumors, with each step in the tumor formation based on the addition of another mutation to the tumor genome. Fig. 137.1A illustrates this concept. The process starts with a single mutation in a single cell. As mutations in critical genes accumulate in a cell, a brain cancer develops in a stepwise fashion, but a tumor does not display malignant features until a critical number of mutations have accumulated. The number of driver mutations that are found in malignant brain tumors ranges from two to about seven, with pediatric brain cancers averaging less.
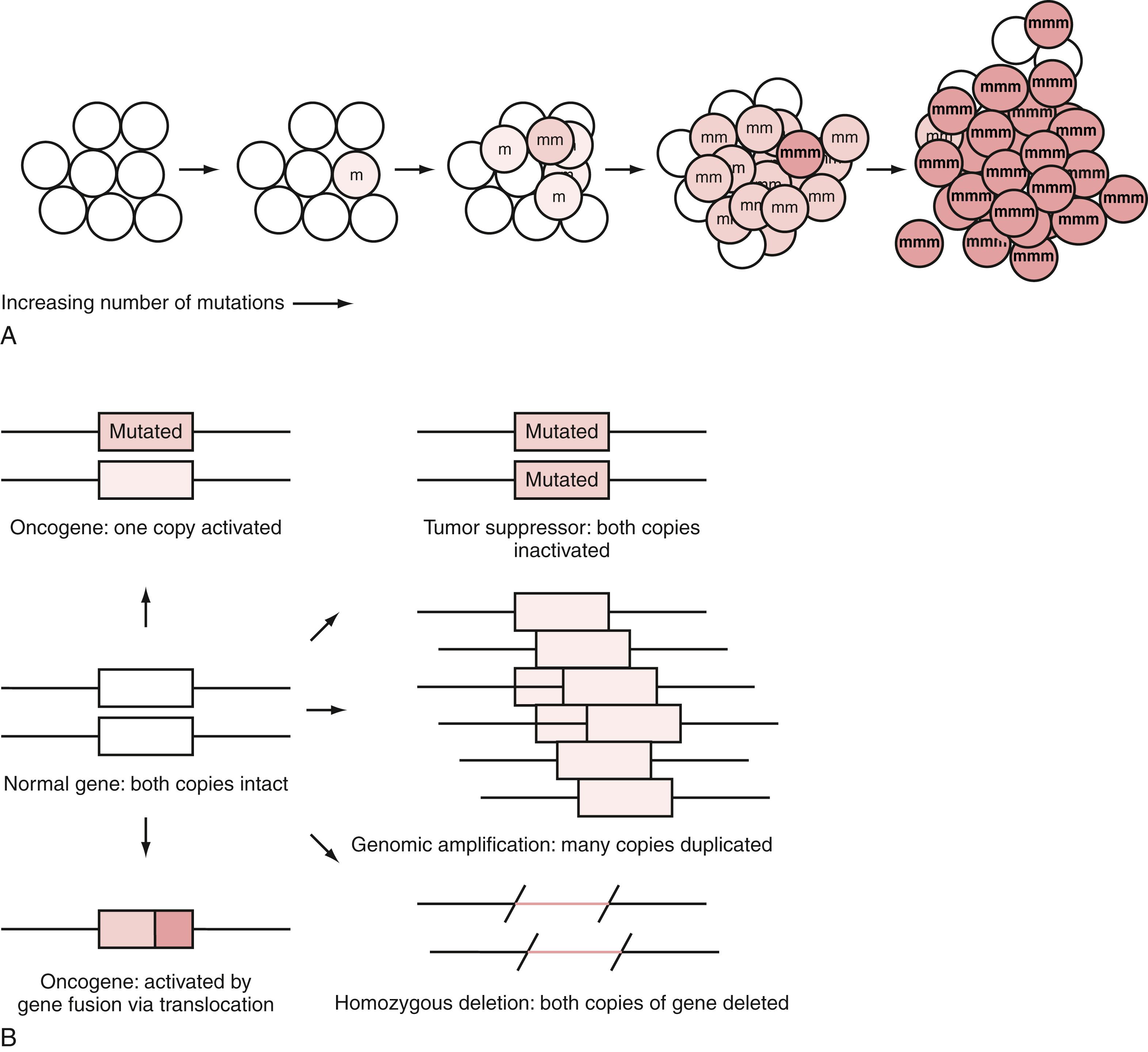
Because clonal expansion likely arises from a single cell, the descendant cells in the tumor should have the same driver mutations necessary to confer a selective advantage. This is found to be the case in cancers, although late-occurring and unstable genomic amplifications give rise to heterogeneity, which is problematic if these amplifications are the target of therapy. For driver mutations, though, the same mutations that initiate and drive cancer formation in its early stages will appear in all parts of the tumor.
Heterogeneity of nondriver mutations is present in brain cancers in the form of diverse low-frequency preexisting mutations in different cells of the tumor. There are enough cells with different mutations from erroneous DNA replication during division that the odds are that there will be at least a few cells that have resistance conveying mutation to any single targeted therapy. The selection pressure of a targeted drug or therapy allows cells with the preexisting resistance mutations to become dominant, rendering the targeted therapy useless for that patient.
Cancer genetics started with the identification of the inherited gene mutations responsible for familial clustering of cancer through linkage or association studies to explain why certain cancers clustered in families. Linkage and association studies can locate the approximate region of the chromosome responsible for a disease phenotype based on finding a polymorphic marker that cosegregates in the families of those affected by the disease. These studies of inheritance of cancer-causing mutations are performed by identifying large families with the disease, collecting blood from family members, and studying candidate markers or markers throughout the genome to find one that is on the same chromosome, close enough to the gene of interest that the marker and mutant gene are not separated by DNA crossing over during meiosis.
The prototypical cancer gene syndrome involves the inheritance of a tumor suppressor mutation wherein there is one mutated allele in the germline, and a second spontaneous mutation inactivates the second allele. Other spontaneous mutations occur subsequently in the cell as part of the cell’s transformation to a malignant state. This first hit dramatically increases the chances of tumor formation in someone with the germline mutation, but by itself is not sufficient.
Because the first hit is inherited as a germline mutation, it can be transmitted to offspring. Although most malignant tumors observed in the clinic do not have an evident hereditary basis, it is important to recognize the possibility of familial clustering of brain tumors and consider genetic counseling and further work-up if evident. Table 137.1 lists some of the more common cancer-associated mendelian disorders that have brain tumors as part of the phenotype. Mendelian disorders are genetic diseases that have a clear pattern of inheritance within families, such as dominant or recessive inheritance. Most of the syndromes that involve brain tumors have an autosomal dominant mendelian inheritance pattern. A complete catalog and literature review on all the mendelian disorders identified is readily accessible at the Online Mendelian Inheritance in Man (OMIM) website.
| Syndrome Name | Alternate Syndrome Name | Gene Symbol(s) | Major Phenotypic Features | Brain Tumors With Increased Risk | References |
|---|---|---|---|---|---|
| Neurofibromatosis type I | Von Recklinghausen disease | NF1 | Café au lait spots and fibromatous skin tumors | Optic pathway glioma, ependymoma, meningiomas, PNET | 9, 52, 53 |
| Mismatch repair cancer syndrome | Turcot syndrome, brain tumor–polyposis (BTP) syndrome type 1 | MLH1, MSH2, MSH6, or PMS | Colorectal cancer without polyposis and glioma | Malignant glioma | 9–11 |
| Li-Fraumeni syndrome | SBLA syndrome | p53, CHEK2, and unknown | Multiple early-onset cancers | Glioma, medulloblastoma, meningioma, others | 9, 54 |
| Adenomatous polyposis of the colon | Gardner syndrome, familial adenomatous polyposis (FAP) | APC | Multiple adenomatous polyps of the colon and rectum wherein one or more likely progress to colon cancer | Medulloblastoma | 9, 55 |
| Basal cell nevus syndrome | Gorlin syndrome | PTCH1 | Basal cell epitheliomas and carcinomas, jaw cysts, and bifid rib | Medulloblastoma | 9, 56 |
| Neurofibromatosis, type 2 | Neurofibromatosis, central type | NF2 | Eighth cranial nerve tumors, meningiomas, and schwannomas | Meningiomas, gliomas, ependymomas | 9, 57 |
| Tuberous sclerosis | Tuberous sclerosis complex | TSC1, TSC2 |
Hamartomas in the brain, skin, heart, kidneys. | Subependymal giant cell astrocytomas | 58 |
In the laboratory the approach used to distinguish a somatic mutation from a hereditary mutation is to sequence the suspected gene mutation in both the tumor and normal tissue. In most cases, lymphocyte DNA from a blood sample is used as the normal control. If the mutation appears only in the tumor, and not in the normal tissue, it is evidence for a somatic mutation acquired during tumor development.
The classic scenario for a brain tumor phenotype with mendelian inheritance is the dominant inheritance of a mutation in a tumor suppressor gene, with an average of 50% of offspring inheriting one copy of the mutated gene from the affected parent. In the case of tumor suppressor genes, both copies need to be inactivated to initiate the process of tumor formation. When one mutation is inherited as the first hit, there is a high probability of the second being inactivated. Inheritance of this first hit greatly increases the risk of cancer.
To find evidence that a DNA sequence is an inherited mutation, DNA samples from affected families are studied using linkage or association studies. If the DNA mutation associates with the affected members in the pedigree(s) significantly more than dictated by chance, than the DNA sequence studied is likely at or near the chromosomal locus responsible for the phenotype. These studies to find mendelian inherited disease are often referred to as linkage studies or identity by descent based on the statistical methods used.
Although most mendelian disorders that include in the phenotype an increased risk for brain tumors are relatively rare in the general population, there are some exceptions. Neurofibromatosis is the most common of the syndromes with brain tumors as a phenotypic feature. Germline mutations in the neurofibromatosis type 1 (NF1) gene result in a syndrome characterized by café au lait spots on the skin, fibromatous tumors, and gliomas and meningiomas. Approximately one person in 4,000 is affected with NF1. About half of the cases are spontaneous and the other half are hereditary. About 10% or less of these patients will develop a malignancy from one of the multiple benign tumors.
The mismatch repair (MMR) cancer syndrome is an important cancer syndrome that involves several different types of cancers, including some glioblastomas, meningiomas, and medulloblastomas. It has an autosomal dominant pattern of inheritance, as do all the syndromes listed in Table 137.1 . There are several DNA repair enzymes that are all necessary for correct DNA MMR in a normal cell. In MMR cancer syndrome, an inactivating mutation in or deletion of MLH1, MSH2, MSH6, or PMS reduces the ability of the cell to identify and correctly repair DNA mismatches that occur during DNA replication. Therefore a mutation in any of these genes can lead to an increased risk for brain and other cancers. The mutations in these “mutator genes” increase the cell’s mutation rate and accelerate the acquisition of mutations elsewhere in the genome, thus leading to a significantly increased risk for developing cancer at an early age.
The adenomatous polyposis coli (APC) syndrome increases risk for colon cancer and brain tumors, as the MMR syndrome does. However, the APC syndrome, or familial adenomatous polyposis (FAP) syndrome, is molecularly and phenotypically distinct from MMR cancer syndrome. This disorder increases risk for medulloblastomas, rather than gliomas, and is due to a mutation in the APC gene. Turcot syndrome was originally applied to any syndrome with colon cancer and brain tumor, but now if used applies only to the MMR cancer syndrome.
Genetic inheritance of cancer risk is of more than scientific interest. Not only do inherited patterns help researchers locate the genes that can initiate brain tumors, but identifying a familial risk of cancer is good clinical practice. A new patient with clustering of brain tumors or other types of tumors in his or her family will often want to know if there is a gene responsible and what the risk is for their family members. Identifying the responsible gene gives the patient options for genetic counseling and for family planning, and provides the opportunity for early diagnosis and treatment of other family members carrying the disease-causing gene allele. In some cases, identifying a cancer syndrome in a family can be lifesaving for a family member.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here