Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Pathology literally translates as the study of suffering (Greek pathos = suffering, logos = study); more prosaically, and as applied to modern medicine, it is the study of disease. Virchow was prescient in asserting that disease originates at the cellular level, but we now appreciate that cellular pathologies arise from perturbations in molecules (genes, proteins, and metabolites) that influence cell survival and behaviors. Thus the foundation of modern pathology is understanding the cellular and molecular aberrations that give rise to diseases. It is illuminating to consider these abnormalities in the context of normal cellular structure and function, which is the subject of this introductory chapter.
It is unrealistic (and even undesirable) to condense the vast and fascinating field of cell biology into a single chapter. Consequently, rather than attempting a comprehensive review, the goal here is to survey basic principles and highlight recent advances that are relevant to the mechanisms of disease that are emphasized throughout the rest of the book.
The sequencing of the human genome at the beginning of the 21st century represented a landmark achievement of biomedical science. Since then the rapidly declining cost of sequencing, the burgeoning computational capacity to mine the ensuing data, and the expanding toolkits to analyze functional outputs (genomics, proteomics, and metabolomics) promise to revolutionize our understanding of health and disease. The emerging information has also revealed a breathtaking level of complexity far beyond the linear sequence of the genome. The potential of these powerful innovations to explain disease pathogenesis and drive therapeutic discovery excites and inspires scientists and the lay public alike.
The human genome contains some 3.2 billion DNA base pairs. Yet, within the genome there are only about 20,000 protein-encoding genes, constituting just 1.5% of the genome. These are the blueprints that instruct the assembly of the enzymes, structural elements, and signaling molecules within the 50 trillion cells that make up the human body. Although 20,000 underestimates the actual number of encoded proteins (many genes produce multiple RNA transcripts that translate to different protein isoforms), it is nevertheless startling to realize that worms, which are composed of fewer than 1000 cells and have 30-fold smaller genomes also have about 20,000 protein-encoding genes. Many of these proteins are recognizable homologs of molecules expressed in humans. What then separates humans from worms?
The answer is not completely known, but evidence suggests that much of the difference lies in the 98.5% of the human genome that does not encode proteins. The function of such long stretches of DNA (so-called genome “dark matter”) was mysterious for many years. However, over 85% of the human genome is ultimately transcribed; nearly 80% is devoted to regulation of gene expression. It follows that while proteins provide the building blocks and machinery required for assembling cells, tissues, and organisms, it is the noncoding regions of the genome that provide the critical “architectural planning.” Practically stated, the difference between worms and humans apparently lies more in the genomic “blueprints” than in the construction materials.
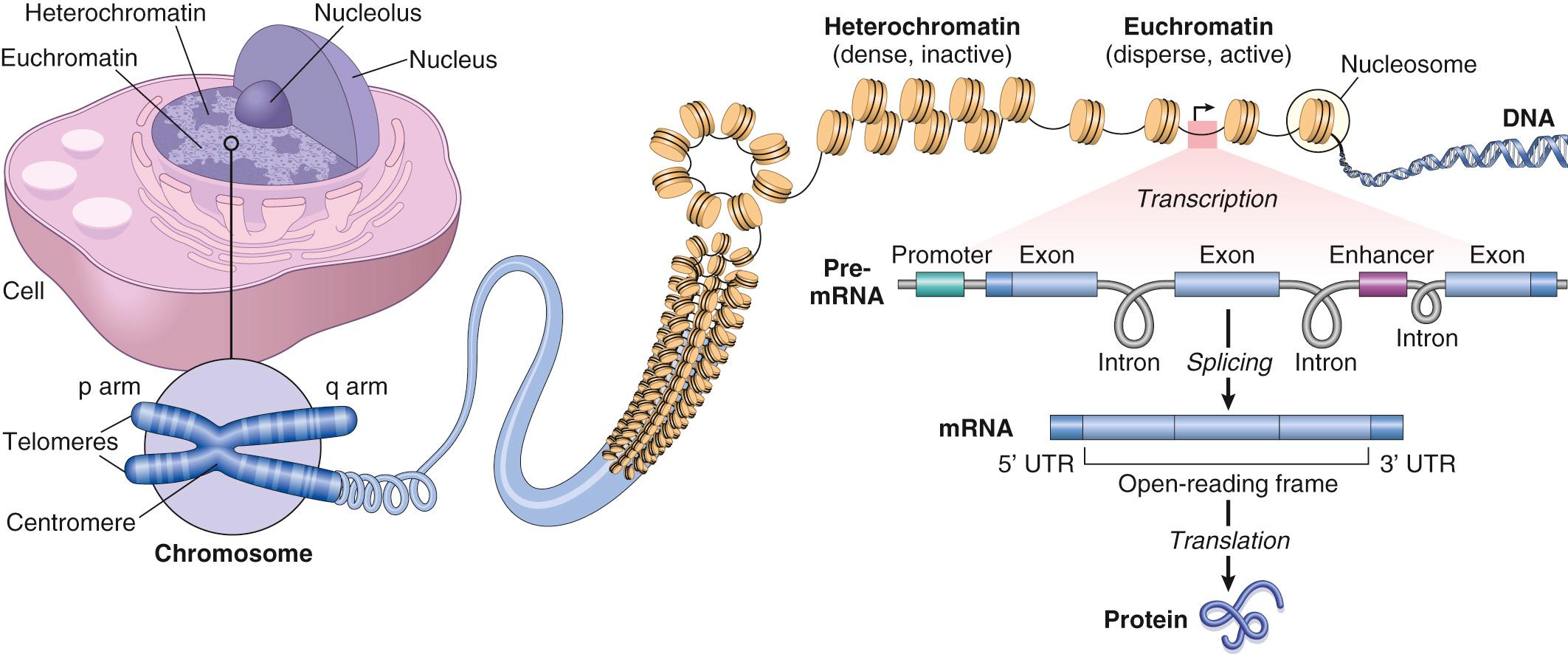
There are five major classes of functional non–protein-coding sequences in the human genome ( Fig. 1.1 ):
Promoter and enhancer regions that provide binding sites for transcription factors.
Binding sites for factors that organize and maintain higher order chromatin structures.
Noncoding regulatory RNAs . Over 60% of the genome is transcribed into RNAs that are never translated but regulate gene expression through a variety of mechanisms. The two best-studied varieties—micro-RNAs (miRNAs) and long noncoding RNAs (lncRNAs)—are described later.
Mobile genetic elements (e.g., transposons ) make up more than a third of the human genome. These “jumping genes” can move around the genome during evolution, resulting in variable copy number and positioning even among closely related species (e.g., humans and other primates). Although implicated in gene regulation and chromatin organization, the function of mobile genetic elements is not well established.
Special structural regions of DNA, in particular, telomeres (chromosome ends) and centromeres (chromosome “tethers”). A major component of centromeres is so-called satellite DNA, consisting of large arrays—up to megabases in length—of repeating sequences (from 5 bp up to 5 kb). Although classically associated with spindle apparatus attachment, satellite DNA is also important in maintaining the dense, tightly packed organization of heterochromatin (discussed later).
Many genetic variations (polymorphisms) associated with diseases are located in non–protein-coding regions of the genome. Thus variation in gene regulation may prove to be more important in disease causation than structural changes in specific proteins. Another surprise that emerged from genome sequencing is that any two humans are typically more than 99.5% DNA-identical (and are 99% sequence-identical with chimpanzees)! Thus individual variation, including differential susceptibility to diseases and environmental stimuli, is encoded in less than 0.5% of our DNA (representing about 15 million bp).
The two most common forms of DNA variation in the human genome are single nucleotide polymorphisms (SNPs) and copy number variations (CNVs).
SNPs are variants at single nucleotide positions and are almost always biallelic (only two choices exist at a given site within the population, such as A or T). Over 6 million human SNPs have been identified, with many showing wide variation in frequency in different populations.
SNPs occur across the genome—within exons, introns, intergenic regions, and coding regions.
Roughly 1% of SNPs occur in coding regions, which is about what would be expected by chance, since coding regions comprise about 1.5% of the genome.
SNPs located in noncoding regions can occur within genomic regulatory elements, thereby altering gene expression; in such instances, SNPs influence disease susceptibility directly.
Some SNPs, termed “neutral” variants, are thought to have no effect on gene function or individual phenotype.
Even “neutral” SNPs may be useful markers if they happen to be coinherited with a disease-associated polymorphism as a result of physical proximity. In other words, the SNP and the causative genetic factor are in linkage disequilibrium.
The effect of most SNPs on disease susceptibility is weak, and it remains to be seen if identification of such variants, alone or in combination, can be used to develop effective strategies to identify those at risk and, ultimately, prevent disease.
CNVs are a form of genetic variation consisting of different numbers of large contiguous stretches of DNA; these can range from 1000 base pairs to millions of base pairs. CNVs can be biallelic and simply duplicated or, alternatively, deleted in some individuals. At other sites there are complex rearrangements of genomic material, with multiple variants in the human population.
CNVs are responsible for between 5 million and 24 million base pairs of sequence difference between any two individuals.
Approximately 50% of CNVs involve gene-coding sequences; thus CNVs may underlie a large portion of human phenotypic diversity.
It is important to note that alterations in DNA sequence cannot by themselves explain the diversity of phenotypes in human populations; moreover, classic genetic inheritance cannot explain differing phenotypes in monozygotic twins. The answers to these conundrums probably lie in epigenetics —heritable changes in gene expression that are not caused by variations in DNA sequence (see the following section).
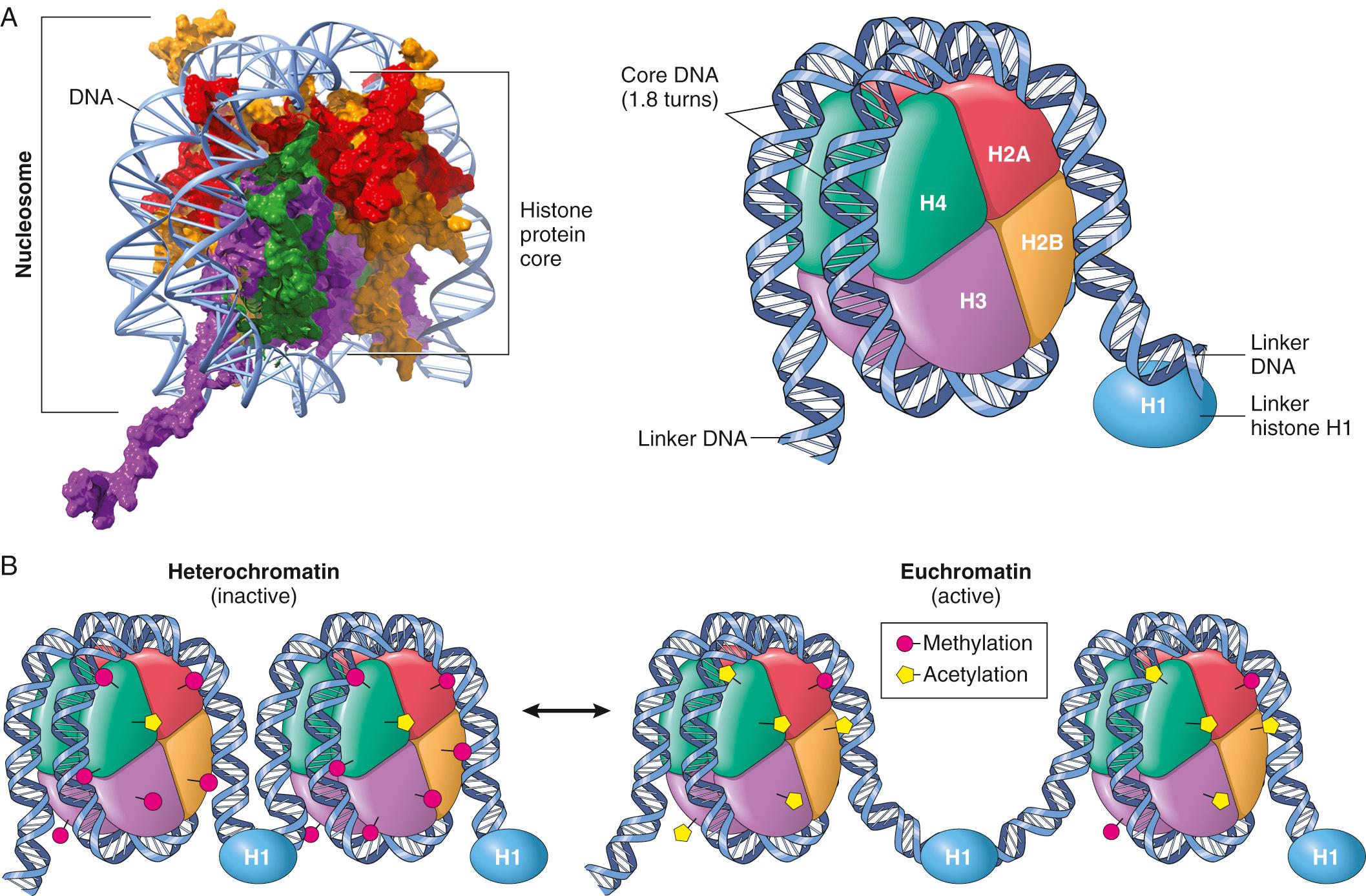
Even though virtually all cells in the body have the same genetic composition, differentiated cells have distinct structures and functions that arise as a result of lineage-specific gene expression programs. Such cell type–specific differences in transcription and translation depend on epigenetic factors (literally, factors that are “above genetics”) that can be conceptualized as follows ( Fig. 1.2 ):
Histones and histone-modifying factors. Nucleosomes consist of DNA segments 147 bp long that are wrapped around a central core structure of highly conserved low molecular weight proteins called histones . The resulting DNA-histone complex resembles a series of beads joined by short DNA linkers. The naked DNA of a single human cell is about 1.8 m long. By winding around histones, like spools of thread, the entire genome can be packed into a nucleus as small as 7 to 8 µm in diameter. In most cases, this structured DNA, termed chromatin, is not wound uniformly. Thus at the light microscopic level, nuclear chromatin is recognizable as cytochemically dense and transcriptionally inactive heterochromatin and disperse, transcriptionally active euchromatin (see Fig. 1.1 ). In general, only the regions that are “unwound” are available for transcription. Chromatin structure can therefore regulate transcription independent of traditional promoters and DNA-binding elements and, due to variations between cell types, helps to define cellular identity and activity.
Histones are not static, but rather are highly dynamic structures regulated by a host of nuclear proteins. Thus chromatin remodeling complexes can reposition nucleosomes on DNA, exposing (or obscuring) gene regulatory elements such as promoters. “Chromatin writer” complexes, on the other hand, carry out over 70 different histone modifications generically denoted as “marks.” Such covalent alterations include methylation, acetylation, or phosphorylation of specific amino acids within histones.
Actively transcribed genes in euchromatin are associated with histone marks that make the DNA accessible to RNA polymerases. In contrast, inactive genes have histone marks that enable DNA compaction into heterochromatin. Histone marks are reversible through the activity of “chromatin erasers.” Still other proteins function as “chromatin readers,” binding histones that bear particular marks and thereby regulating gene expression.
Histone methylation. Both lysines and arginines can be methylated by specific writer enzymes; methylation of histone lysine residues can lead to transcriptional activation or repression, depending on which histone residue is marked.
Histone acetylation. Lysine residues are acetylated by histone acetyltransferases (HATs), whose modifications tend to open the chromatin and increase transcription. In turn, these changes can be reversed by histone deacetylases (HDACs), leading to chromatin condensation.
Histone phosphorylation. Serine residues can be modified by phosphorylation; depending on the specific residue, the DNA may be opened for transcription or condensed and inactive.
DNA methylation. High levels of DNA methylation in gene regulatory elements typically result in transcriptional silencing. Like histone modifications, DNA methylation is tightly regulated by methyltransferases, demethylating enzymes, and methylated-DNA-binding proteins.
Chromatin organizing factors. Much less is known about these proteins, which are believed to bind to noncoding regions and control long-range looping of DNA, thus regulating the spatial relationships between enhancers and promoters that control gene expression.
Deciphering the mechanisms that allow epigenetic factors to control genomic organization and gene expression in a cell-type-specific fashion is an extraordinarily complex proposition. Despite the intricacies, there is already ample evidence that dysregulation of the “epigenome” has a central role in malignancy ( Chapter 7 ), and emerging data indicate that many other diseases are associated with inherited or acquired epigenetic alterations. Unlike genetic changes, many epigenetic alterations (e.g., histone acetylation and DNA methylation) are reversible and amenable to therapeutic intervention; HDAC and DNA methylation inhibitors are already being tested in the treatment of various forms of cancer.
Genes can also be regulated by noncoding RNAs. These genomic sequences are transcribed but not translated. Although many distinct families of noncoding RNAs exist, we will discuss only two examples here: small RNA molecules called microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) (>200 nucleotides in length).
miRNAs do not encode proteins; they modulate translation of target messenger RNAs (mRNAs). Posttranscriptional silencing of gene expression by miRNA is a fundamental and well-conserved mechanism of gene regulation present in all eukaryotes (plants, animals, and fungi). Even bacteria have a primitive version of the same machinery that they use to protect themselves against foreign DNA (e.g., phage and virus DNA). The profound influence of miRNAs on protein expression allows these relatively short RNAs (22 nucleotides on average) to be critical regulators of developmental pathways as well as pathologic conditions (e.g., cancer).
The human genome encodes almost 6000 miRNA genes, about 30% of the total number of protein-coding genes. Individual miRNAs can regulate multiple protein-coding genes, allowing each miRNA to coregulate entire programs of gene expression. Transcription of miRNA genes produces a primary transcript (pri-miRNA) that is processed into progressively smaller segments, including trimming by the enzyme Dicer. This generates mature single-stranded miRNAs of 21 to 30 nucleotides that associate with a multiprotein aggregate called RNA-induced silencing complex (RISC) ( Fig. 1.3 ). Subsequent base pairing between the miRNA strand and its target mRNA directs the RISC to either induce mRNA cleavage or repress its translation. In this way the target mRNA is posttranscriptionally silenced .
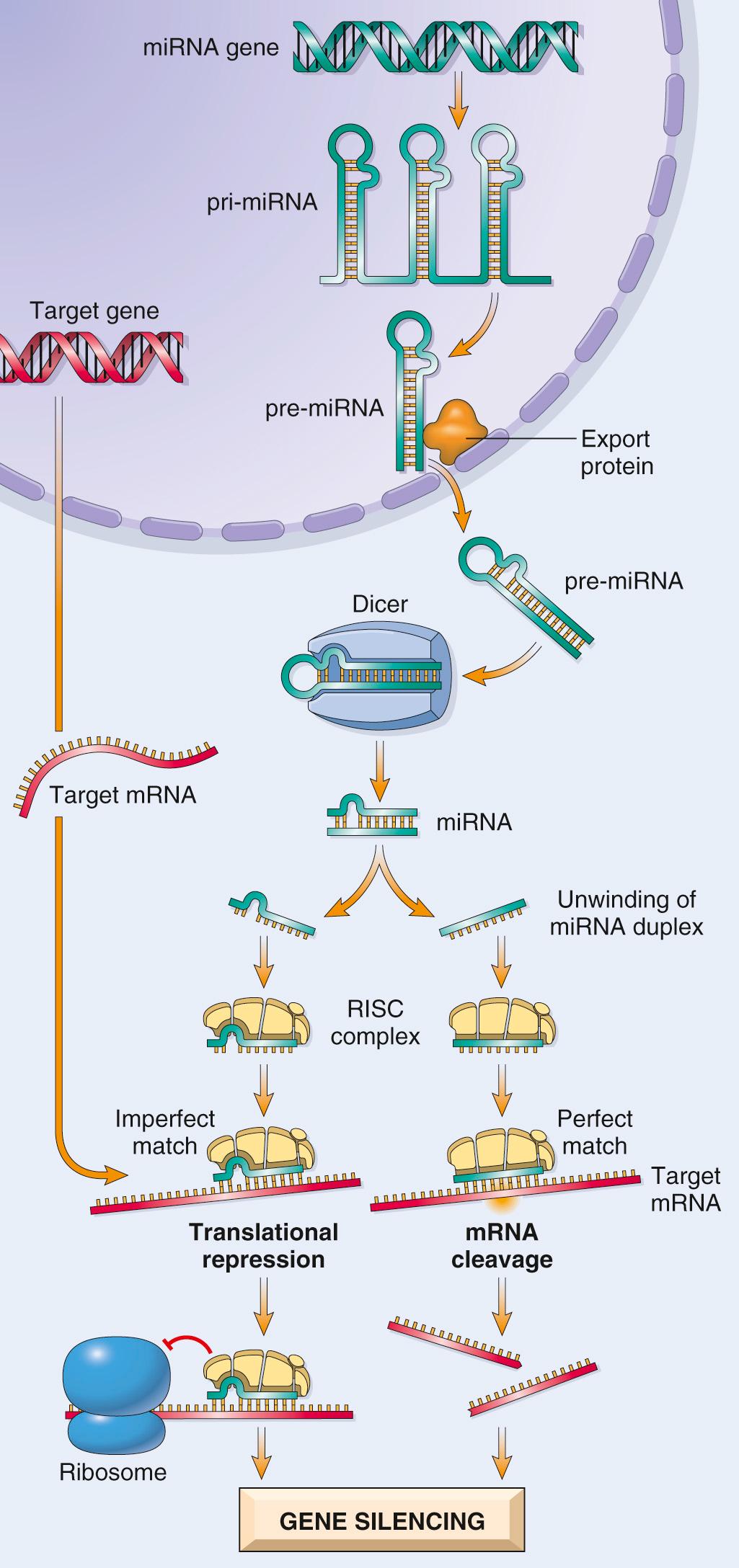
Small interfering RNAs (siRNAs) are short RNA sequences that can be introduced experimentally into cells where they serve as substrates for Dicer and interact with RISC, thereby reproducing endogenous miRNAs function. Synthetic siRNAs that target specific mRNA species are powerful laboratory tools to study gene function (so-called knockdown technology) and are also being studied as potential therapeutic agents to silence pathogenic genes (e.g., oncogenes that drive neoplastic transformation).
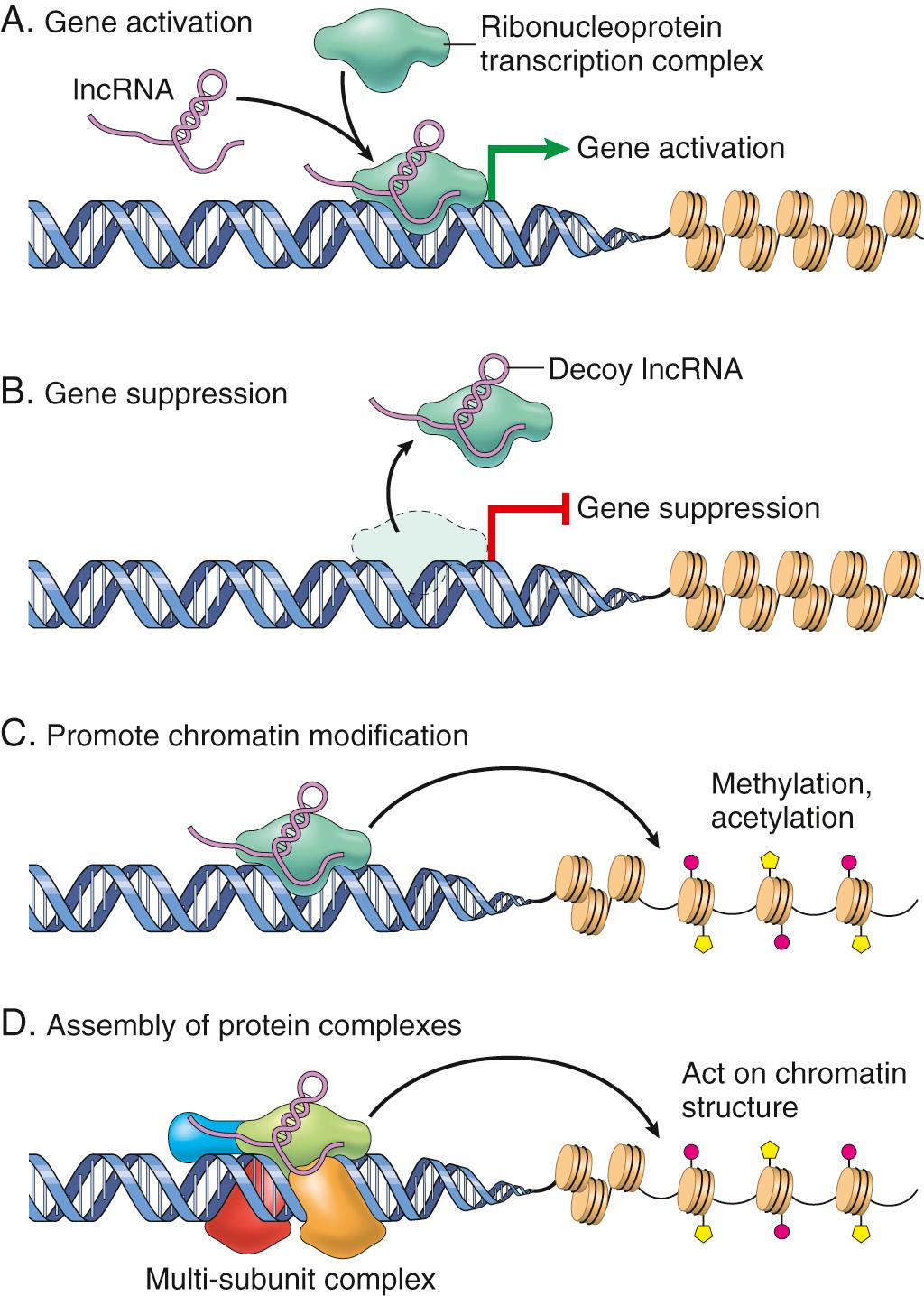
Recent studies have further identified an untapped universe of lncRNAs—by some calculations, the number of lncRNAs may exceed coding mRNAs by 10-fold to 20-fold. lncRNAs modulate gene expression by several mechanisms ( Fig. 1.4 ). As one example, lncRNAs can bind to chromatin and restrict RNA polymerase from accessing coding genes within that region. The best-known example is XIST, which is transcribed from the X chromosome and plays an essential role in the physiologic X chromosome inactivation that occurs in females. XIST itself escapes X inactivation but forms a repressive “cloak” on the X chromosome from which it is transcribed, resulting in gene silencing. Conversely, many enhancers are actually sites of lncRNA synthesis. In this case the lncRNAs expand transcription from gene promoters via a variety of mechanisms (see Fig. 1.4 ).
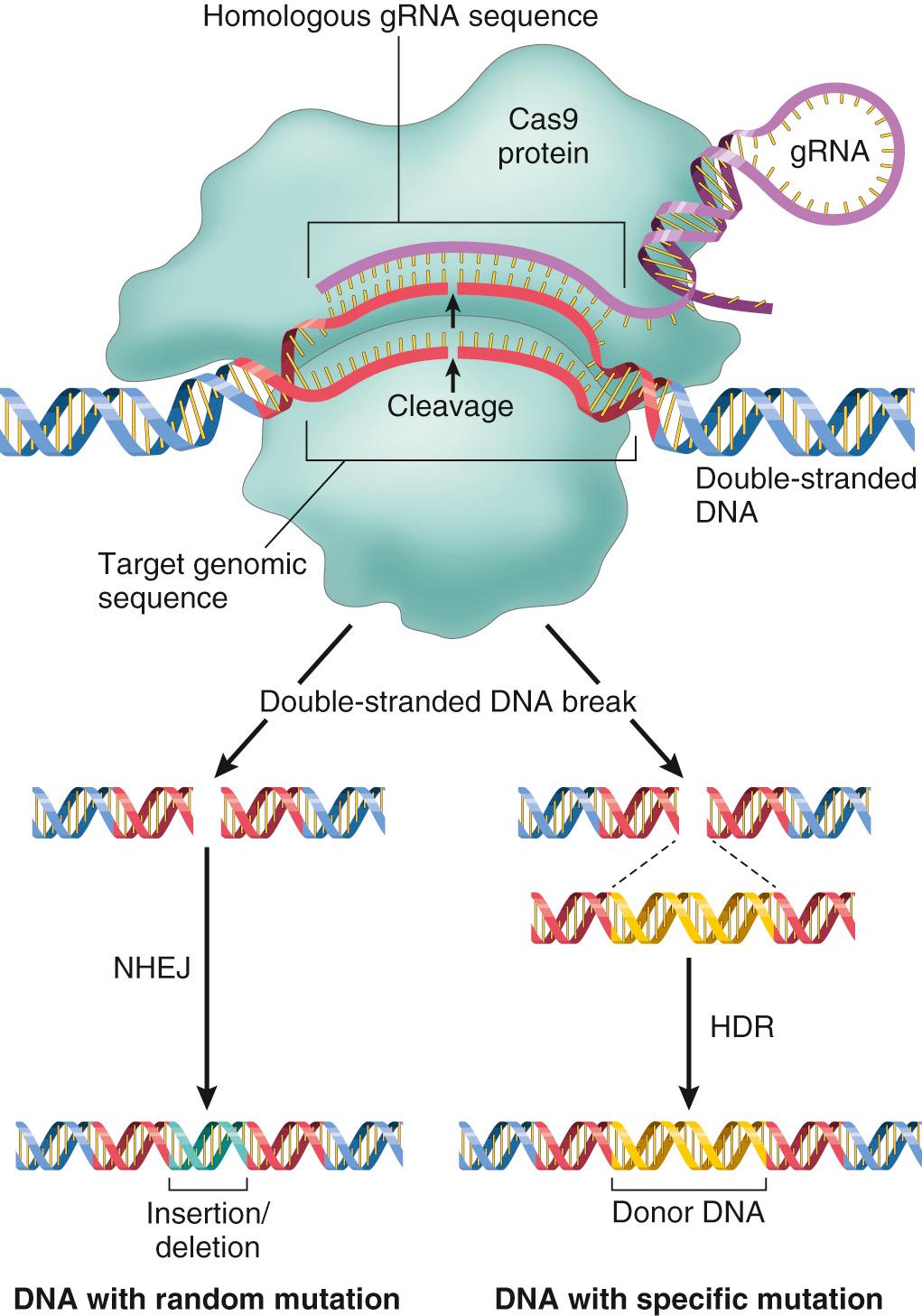
An exciting new development that allows high-fidelity genome editing may usher in the next era of the molecular revolution. This advance comes from a wholly unexpected source: the discovery of clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated genes (Cas), such as the Cas9 nuclease. These are linked genetic elements that endow prokaryotes with a form of acquired immunity to phages and plasmids. Bacteria use the system to sample the DNA of infecting agents and integrate portions into their genomes as CRISPRs. These CRISPR segments are subsequently transcribed and processed into guide RNA sequences that bind and direct the Cas9 nuclease to specific sites (e.g., a phage sequence) so that it can be cleaved to disable the infecting agent.
Gene editing repurposes this process by using artificial 20-base guide RNAs (gRNAs) that bind Cas9 and are complementary to a targeted DNA sequence ( Fig. 1.5 ). Cas9 then induces double-stranded DNA breaks at the site of gRNA binding. Repair of the highly specific cleavages can lead to random disruptive mutations (through nonhomologous end joining) or can introduce new genetic material with precision (by homologous recombination). Both the guide sequences and the Cas enzyme, either as a coding DNA (cDNA) or a protein, can be easily introduced into cells. The potential application to genetic engineering, due to the impressive specificity of the Cas9 system (up to 10,000-fold better than other previous editing systems), has led to great excitement. Applications include inserting specific mutations in cells and tissues to model cancers and other diseases and rapidly generating transgenic animal models from edited embryonic stem cells. CRISPR also makes it possible to selectively edit mutations that cause hereditable disease, or—perhaps more worrisome—to just eliminate less “desirable” traits. Predictably the technology has inspired a vigorous debate regarding the ethics of its use.
Normal functioning and intracellular homeostasis depend on a variety of fundamental cell housekeeping functions that all differentiated cells must perform to maintain viability and normal activity. These include protection from the environment, nutrient acquisition, metabolism, communication, movement, renewal of senescent molecules, molecular catabolism, and energy generation.
Many of the normal housekeeping functions of the cell are compartmentalized within membrane bound intracellular organelles ( Fig. 1.6 ). By isolating certain cellular functions within distinct compartments, potentially injurious degradative enzymes or toxic metabolites can be kept at usefully high concentrations without risking damage to more delicate intracellular constituents. Moreover, compartmentalization also allows the creation of unique intracellular environments (e.g., low pH or high calcium) that permit more efficient functioning of certain enzymes or metabolic pathways.
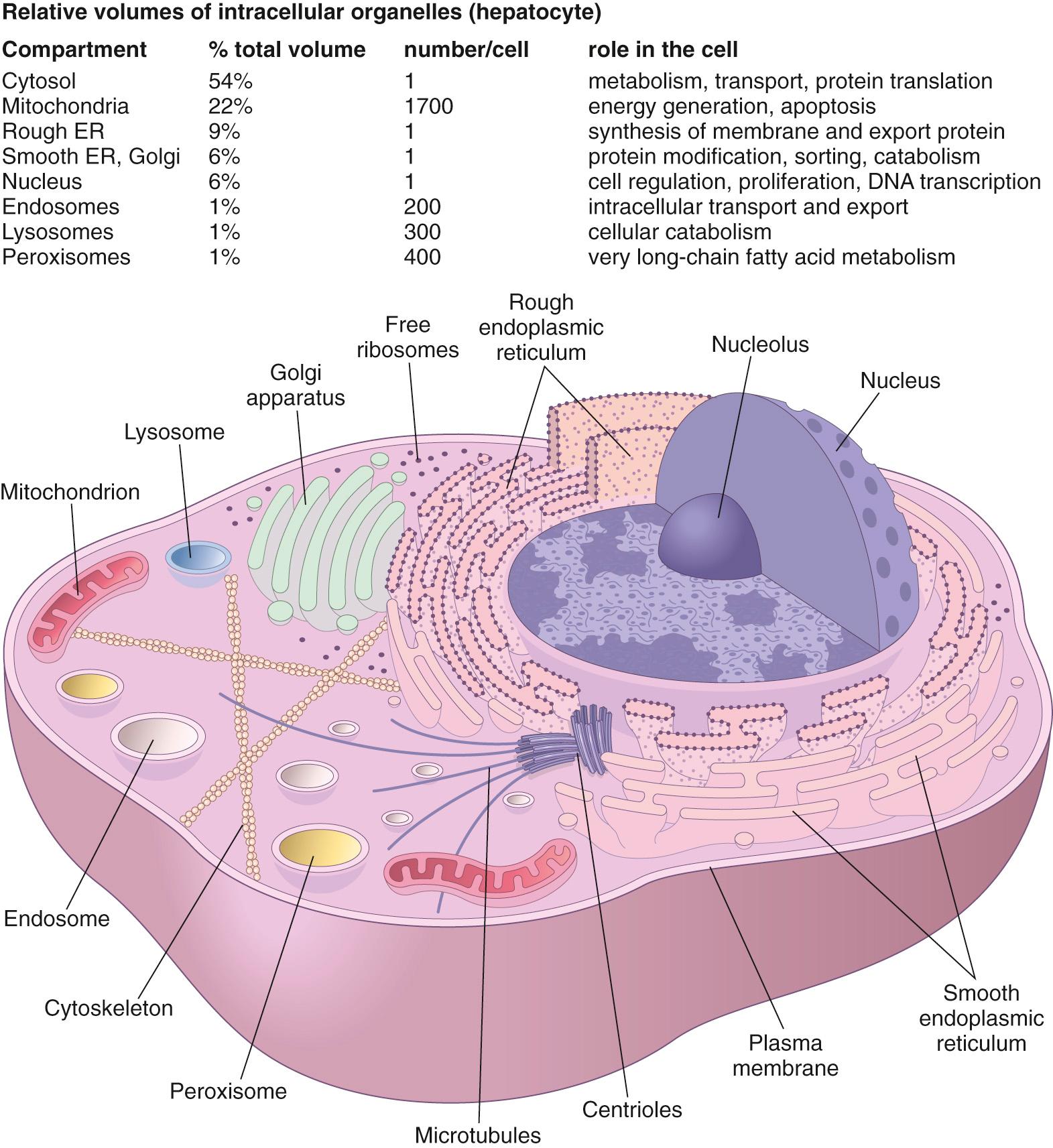
New proteins destined for the plasma membrane or secretion are physically assembled in the rough endoplasmic reticulum (RER) and Golgi apparatus; proteins intended for the cytosol are synthesized on free ribosomes. Smooth endoplasmic reticulum (SER) is used for steroid hormone and lipoprotein synthesis and modification of hydrophobic compounds into water-soluble molecules for export.
Cells catabolize the wide variety of molecules that they endocytose, as well as the entire repertoire of their own proteins and organelles—all of which are constantly being degraded and renewed. Breakdown of these constituents takes place at three different sites, ultimately serving different functions.
Proteasomes are “disposal” complexes that degrade denatured or otherwise “tagged” cytosolic proteins. In antigen-presenting cells, the resulting short peptides are presented in the context of class I or class II major histocompatibility molecules to help drive the adaptive immune response ( Chapter 6 ). In other cases, proteasomal degradation of regulatory proteins or transcription factors can trigger initiation or suppression of signaling pathways.
Lysosomes are intracellular organelles containing degradative enzymes that permit digestion of a wide range of macromolecules, including proteins, polysaccharides, lipids, and nucleic acids. They are the site of senescent intracellular organelle breakdown (a process called autophagy ) and where phagocytosed microbes are killed and catabolized.
Peroxisomes contain catalase, peroxidase, and other oxidative enzymes; they play a specialized role in the breakdown of very long-chain fatty acids, generating hydrogen peroxide in the process.
The contents and location of cellular organelles are also highly regulated. Endosomal vesicles shuttle internalized material to the appropriate intracellular site(s), while other membrane-bound vesicles direct newly synthesized materials to the cell surface or specific organelles. Movement—of both organelles and proteins within the cell, as well as the entire cell in its environment—is accomplished by the cytoskeleton, which is composed of filamentous actin (microfilaments), keratins (intermediate filaments), and microtubules. These structural proteins also maintain cellular shape and intracellular organization, which are essential to generation and maintenance of cell polarity. This is particularly important in epithelium where the top of the cell (apical) and the bottom and sides of the cell (basolateral) are exposed to different environments and have distinct functions. Loss of polarity could, for example, disrupt vectorial transcellular transport in the intestine or renal tubule.
Cell growth and maintenance require a constant supply of both energy and the building blocks that are needed for synthesis of macromolecules. Most of the adenosine triphosphate (ATP) that powers cells is generated via mitochondrial oxidative phosphorylation. Mitochondria also serve as an important source of metabolic intermediates needed for anabolic metabolism, are sites of synthesis of certain macromolecules (e.g., heme), and contain important sensors of cell damage that can initiate and regulate programmed cell death (e.g., apoptosis).
In growing and dividing cells, all of these organelles have to be replicated (organellar biogenesis) and correctly apportioned in daughter cells following mitosis. Moreover, because the macromolecules and organelles have finite lifespans (mitochondria, for example, last only about 10 days), mechanisms must also exist that allow for the recognition and degradation of “worn-out” cellular components.
With this as a primer, we will now move on to discuss cellular components and their function in greater detail.
Plasma membranes (and all other organellar membranes for that matter) are more than just static lipid sheaths. Rather, they are fluid bilayers of amphipathic phospholipids—hydrophilic head groups that face the aqueous environment and hydrophobic lipid tails that interact with each other to form a barrier to passive diffusion of large or charged molecules ( Fig. 1.7 ). The bilayer has a remarkably heterogeneous composition of different phospholipids that vary by location and are also asymmetric—that is, membrane lipids preferentially associate with extracellular or cytosolic faces. Proper localization of these molecules is important for cell health. For example, specific phospholipids interact with particular membrane proteins and modify their distributions and functions.
Phosphatidylinositol on the inner membrane leaflet can be phosphorylated, serving as an electrostatic scaffold for intracellular proteins; alternatively, polyphosphoinositides can be hydrolyzed by phospholipase C to generate intracellular second signals like diacylglycerol and inositol trisphosphate.
Phosphatidylserine is normally restricted to the inner face where it confers a negative charge involved in electrostatic protein interactions; however, when flipped to the extracellular leaflet, it becomes a potent “eat me” signal during programmed cell death (e.g., apoptosis). In platelets, phosphatidylserine is also a cofactor in blood clotting.
Glycolipids and sphingomyelin are preferentially located on the extracellular face; glycolipids, including gangliosides with complex sugar linkages and terminal sialic acids that confer negative charges, support charge-based interactions that contribute to including inflammatory cell recruitment and sperm-egg fusion.
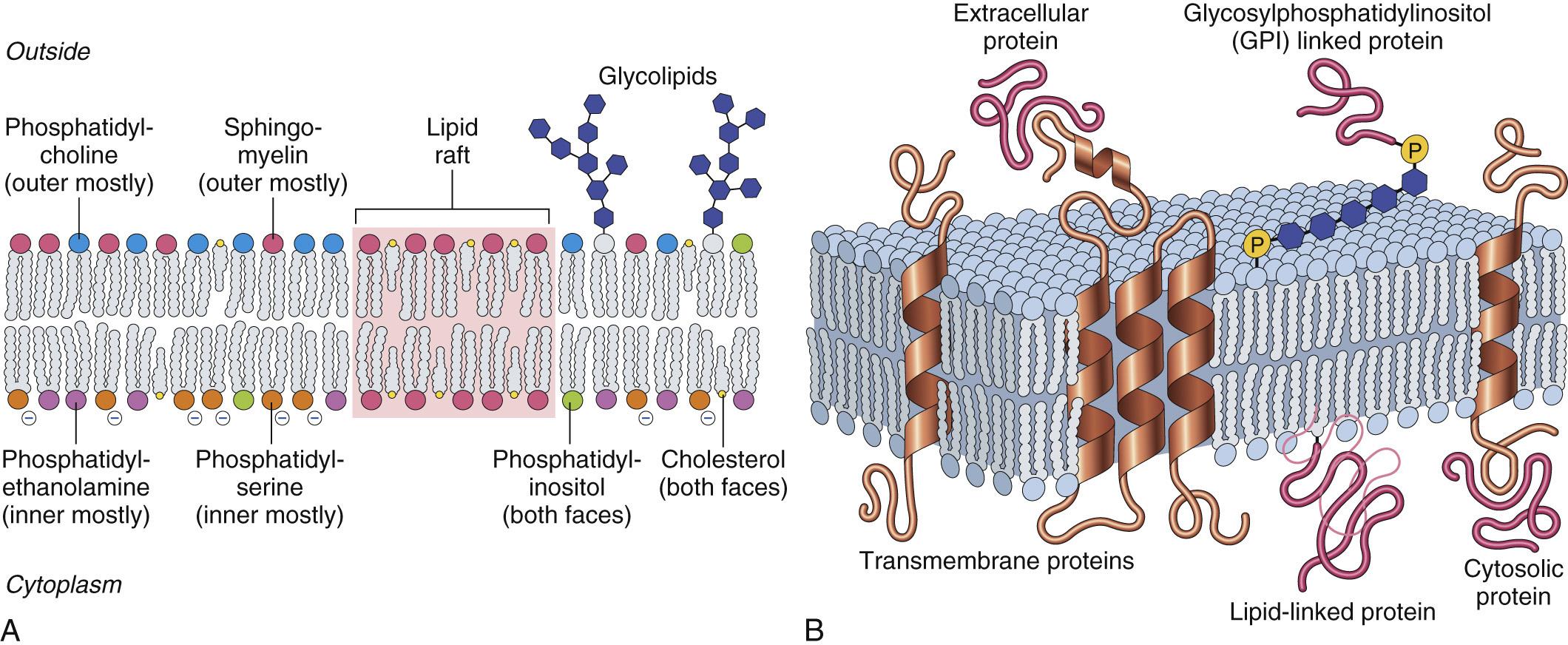
Despite substantial lateral fluidity, some membrane constituents concentrate into specialized domains (e.g., lipid rafts ) that are enriched in glycosphingolipids and cholesterol. Since inserted membrane proteins have different intrinsic solubilities in domains with distinct lipid compositions, this membrane organization also impacts protein distribution. This geographic organization of plasma membrane components impacts cell-cell and cell-matrix interactions, intracellular signaling, and the specialized sites of vesicle budding or fusion.
The plasma membrane is liberally studded with a variety of proteins and glycoproteins involved in (1) ion and metabolite transport; (2) fluid-phase and receptor-mediated uptake of macromolecules; and (3) cell-ligand, cell-matrix, and cell-cell interactions. The means by which these proteins associate with membranes frequently reflects function. For example, multiple transmembrane-spanning proteins are often pores or molecular transporters, while proteins that are superficially attached to the membrane via labile linkages are more likely to participate in signaling. In general, proteins associate with the lipid bilayer by one of four mechanisms.
Most proteins are integral or transmembrane proteins, having one or more relatively hydrophobic α-helical segments that traverse the lipid bilayer.
Proteins synthesized on free ribosomes in the cytosol may be modified posttranslationally by addition of prenyl groups (e.g., farnesyl, related to cholesterol) or fatty acids (e.g., palmitic or myristic acid) that insert into the cytosolic side of the plasma membrane.
Proteins on the extracellular face of the membrane may be anchored by glycosylphosphatidylinositol (GPI) tails that are added posttranslationally.
Peripheral membrane proteins may noncovalently associate with true transmembrane proteins.
Many plasma membrane proteins function as large complexes; these may be aggregated either under the control of chaperone molecules in the RER or by lateral diffusion in the plasma membrane, followed by complex formation in situ. For example, many protein receptors (e.g., cytokine receptors) dimerize or trimerize in the presence of ligand to form functional signaling units. Although lipid bilayers are fluid within the plane of the membrane, components can be confined to discrete domains. This can occur by localization to lipid rafts (discussed earlier) or through intercellular protein-protein interactions (e.g., tight junctions ) that establish discrete boundaries and also have unique lipid composition. The latter strategy is used to maintain cell polarity (e.g., top/apical/free vs. bottom/basolateral/bound to extracellular matrix [ECM]) in epithelial cells. Interactions of other membrane and cytosolic proteins with one another and the cytoskeleton also contributes to cell polarity.
The extracellular face of the plasma membrane is diffusely decorated by carbohydrates, not only as complex oligosaccharides on glycoproteins and glycolipids, but also as polysaccharide chains attached to integral membrane proteoglycans. This glycocalyx can form a chemical and mechanical barrier.
Although the barrier provided by plasma membranes is critical, transport of selected molecules across the lipid bilayer or to intracellular sites via vesicular transport is essential. Several mechanisms contribute to this transport.
Small, nonpolar molecules like O 2 and CO 2 readily dissolve in lipid bilayers and therefore rapidly diffuse across them. Larger hydrophobic molecules, (e.g., steroid-based molecules like estradiol or vitamin D) can also cross lipid bilayers with relative impunity. While small polar molecules such as water (18 Da) can also diffuse across membranes at low rates, in tissues responsible for significant water movement (e.g., renal tubular epithelium), special integral membrane proteins called aquaporins form transmembrane channels for water, H 2 O 2 , and other small molecules. In contrast, the lipid bilayer is an effective barrier to the passage of larger polar molecules (>75 Da); at 180 Da, for example, glucose is effectively excluded. Lipid bilayers are also impermeant to ions due to their charge and hydration.
Plasma membrane transport proteins are required for uptake and secretion of ions and larger molecules that are required for cellular function (e.g., nutrient uptake and waste disposal). Ions and small molecules can be transported by channel proteins and carrier proteins. Similar pores and channels also mediate transport across organellar membranes. These transporters that move ions, sugars, nucleotides, etc., frequently have exquisite specificities, and can be either active or passive (see below). For example, some transporters accommodate glucose but reject galactose.
Channel proteins create hydrophilic pores, which, when open, permit rapid movement of solutes (usually restricted by size and charge).
Carrier proteins bind their specific solute and undergo a series of conformational changes to transfer the ligand across the membrane; their transport is relatively slow.
Solute transport across the plasma membrane is frequently driven by a concentration and/or electrical gradient between the inside and outside of the cell via passive transport (virtually all plasma membranes have an electrical potential difference across them, with the inside negative relative to the outside). In other cases, active transport of certain solutes (against a concentration gradient) is accomplished by carrier molecules (never channels) at the expense of ATP hydrolysis or a coupled ion gradient. For example, most apical nutrient transporters in the intestines and renal tubules exploit the extracellular to intracellular Na + gradient to allow absorption even when intracellular nutrient concentrations exceed extracellular concentrations. This form of active transport does not use ATP directly, but depends on the Na + gradient generated by Na + -Ka + ATPase. Other transporters are ATPases. One example is the multidrug resistance (MDR) protein , which pumps polar compounds (e.g., chemotherapeutic drugs) out of cells and may render cancer cells resistant to treatment.
Water movement into or out of cells is passive and directed by solute concentrations. Thus extracellular salt in excess of that in the cytoplasm (hypertonicity) causes net movement of water out of cells, while hypotonicity causes net movement of water into cells. Conversely, the charged metabolites and proteins within the cytoplasm attract charged counterions that increase intracellular osmolarity. Thus to prevent overhydration, cells must constantly pump out small inorganic ions (e.g., Na + )—typically through the activity of the membrane ion-exchanging ATPase. Loss of the ability to generate energy (e.g., in a cell injured by toxins or ischemia) therefore results in osmotic swelling and eventual cell rupture. Similar transport mechanisms also regulate concentrations of other ions (e.g., Ca 2+ and H + ). This is critical to many processes. For example, cytosolic enzymes are most active at pH 7.4 and are often regulated by Ca 2+ , whereas lysosomal enzymes function best at pH 5 or less.
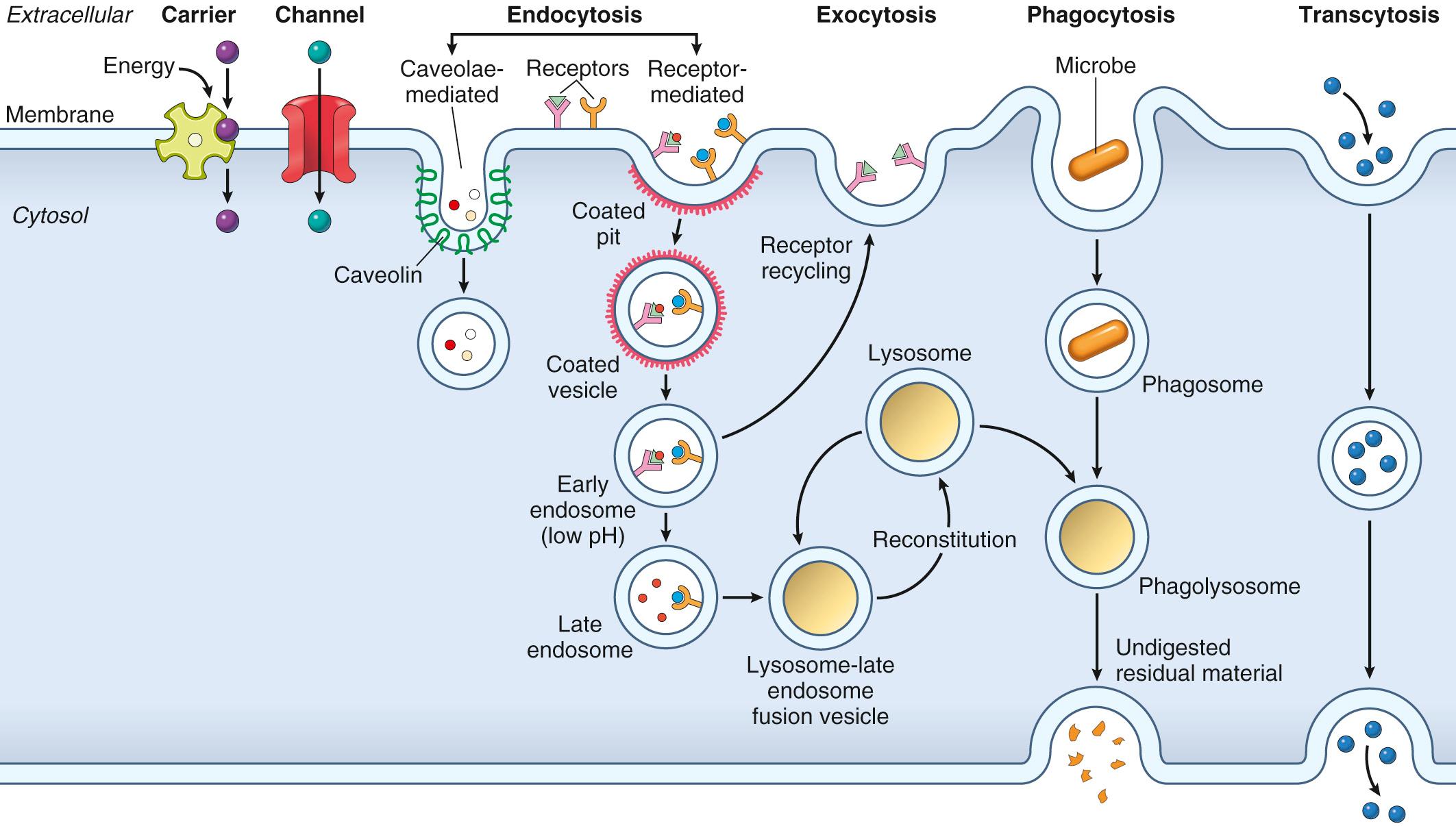
Uptake of fluids or macromolecules by the cell is called endocytosis . Depending on the size of the vesicle, endocytosis may be denoted pinocytosis (“cellular drinking”) or phagocytosis (“cellular eating”). Generally, phagocytosis is restricted to certain cell types (e.g., macrophages and neutrophils) whose role is to specifically ingest invading organisms or dead cell fragments.
Certain small molecules—including some vitamins—bind to cell-surface receptors and are taken up through invaginations of the plasma membrane called caveolae. Uptake can also occur through membrane invaginations coated by an intracellular matrix of clathrin proteins that spontaneously assemble into a basket-like lattice which helps drive endocytosis (discussed more later). In both cases, activity of the “pinchase” dynamin is required for vesicle release.
Macromolecules can also be exported from cells by exocytosis. In this process, proteins synthesized and packaged within the RER and Golgi apparatus are concentrated in secretory vesicles, which then fuse with the plasma membrane to expel their contents. Common examples include peptide hormones (e.g., insulin) and cytokines.
Transcytosis is the movement of endocytosed vesicles between the apical and basolateral compartments of cells. This is a mechanism for transferring large amounts of intact proteins across epithelial barriers (e.g., ingested antibodies in maternal milk) or for rapid movement of large solute volumes.
We now return to the specifics of endocytosis (see Fig. 1.8 ).
Caveolae-mediated endocytosis. Caveolae (“little caves”) are noncoated plasma membrane invaginations associated with GPI-linked molecules, cyclic adenosine monophosphate (cAMP) binding proteins, src-family kinases, and the folate receptor; caveolin is the major structural protein of caveolae, which, like membrane rafts (see above), are enriched in glycosphingolipids and cholesterol. Internalization of caveolae along with bound molecules and associated extracellular fluid is called potocytosis —literally “cellular sipping.” In addition to supporting transmembrane delivery of some molecules (e.g., folate), caveolae regulate transmembrane signaling and cellular adhesion via internalization of receptors and integrins.
Receptor-mediated endocytosis. Macromolecules bound to membrane receptors (such as transferrin or low-density lipoprotein [LDL] receptors) are taken up at specialized regions of the plasma membrane called clathrin-coated pits . The receptors are efficiently internalized by membrane invaginations driven by the associated clathrin matrix, eventually pinching off to form clathrin-coated vesicles . Trapped within these vesicles is also a gulp of the extracellular milieu (fluid-phase pinocytosis). The vesicles then rapidly lose their clathrin coating and fuse with an acidic intracellular structure called the early endosome; the endosomal vesicles undergo progressive maturation to late endosomes, ultimately fusing with lysosomes. In the acidic environment of the endosomes, LDL and transferrin receptors release their cargo (cholesterol and iron, respectively), which is then transported into the cytosol.
After release of bound ligand, some receptors recycle to the plasma membrane and are reused (e.g., transferrin and LDL receptors), while others are degraded within lysosomes (e.g., epidermal growth factor receptor). In the latter case, degradation after internalization results in receptor downregulation that limits receptor-mediated signaling. Defects in receptor-mediated transport of LDL underlie familial hypercholesterolemia, as described in Chapter 5 .
Endocytosis requires recycling of internalized vesicles back to the plasma membrane (exocytosis) for another round of ingestion. This is critical, as a cell will typically ingest from the extracellular space the equivalent of 10% to 20% of its own cell volume each hour—amounting to 1% to 2% of its plasma membrane each minute! Without recycling, the plasma membrane would be rapidly depleted. Endocytosis and exocytosis must therefore be tightly coupled to avoid large changes in plasma membrane area.
The ability of cells to adopt a particular shape, maintain polarity, organize intracellular organelles, and migrate depends on an intracellular scaffold of structural proteins that form the cytoskeleton ( Fig. 1.9 ). Although the cytoskeleton can have an appearance similar to the beams and supports of a building, cytoskeletal structures are constantly elongating and shrinking; microfilaments and microtubules are most active, and their assembly and disassembly can drive cell migration.
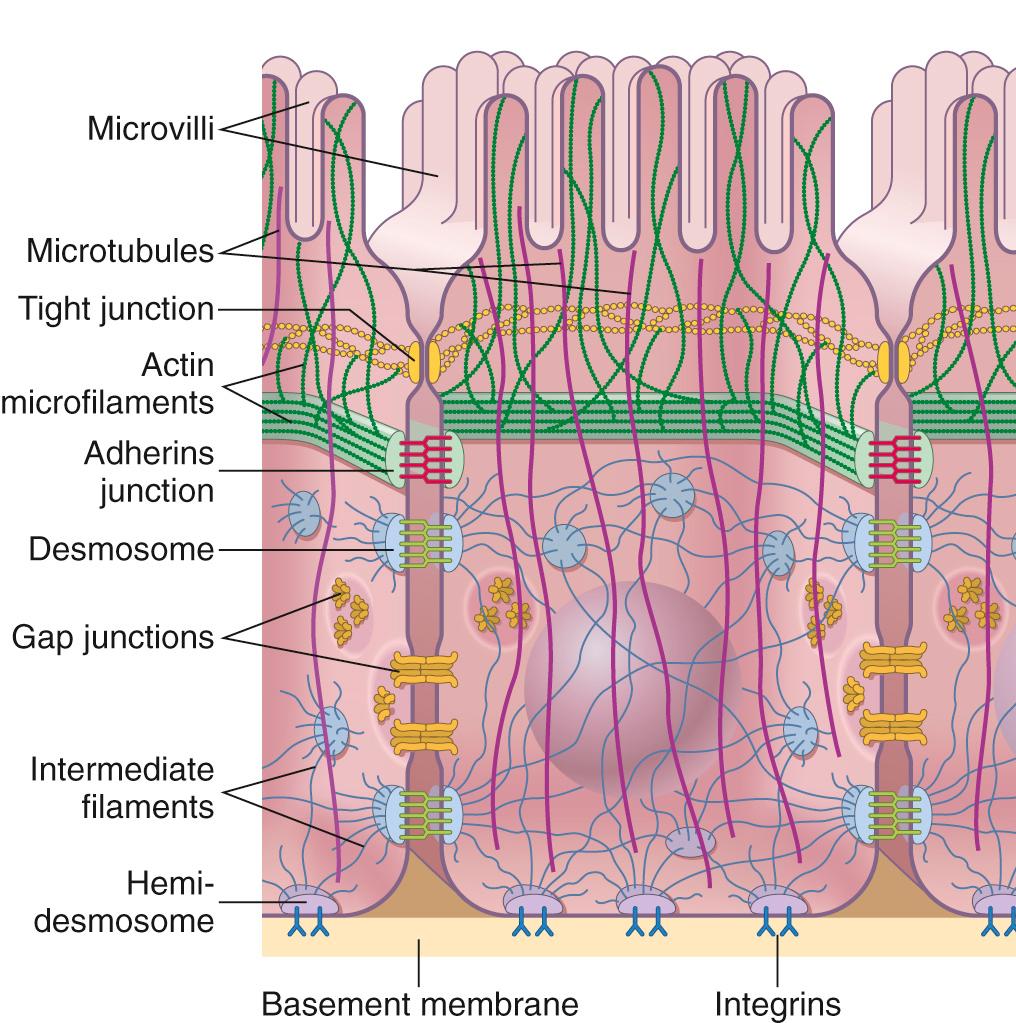
In eukaryotic cells, there are three major classes of cytoskeletal proteins.
Actin microfilaments are 5- to 9-nm diameter fibrils formed from the globular protein actin (G-actin), the most abundant cytosolic protein in cells. G-actin monomers noncovalently polymerize into long filaments (F-actin) that intertwine to form double-stranded helices with a defined polarity. Although the details are (as always) more nuanced, new subunits are typically added at the “positive” end of the strand and removed from the “negative” end—a process referred to as actin treadmilling . Actin nucleating, binding, and regulatory proteins organize polymerization, bundling, and branching to form networks that control cell shape and movement. This complex and its association with motor proteins (e.g., myosin) is so precisely arrayed in skeletal and cardiac muscle that a banding pattern is apparent by light microscopy. ATP hydrolysis by myosin slides the actin filaments relative to one another to cause muscle contraction. Although less coordinated, myosins, of which there are many, are responsible for other functions that depend on actin contraction including vesicular transport, epithelial barrier regulation, and cell migration.
Intermediate filaments are 10-nm diameter fibrils that comprise a large and heterogeneous family that includes keratin proteins and nuclear lamins. Intermediate filaments predominantly form ropelike polymers and do not usually actively reorganize like actin and microtubules. This allows intermediate filaments to provide tensile strength so that cells can bear mechanical stress, e.g., in epithelia where intermediate filaments link desmosomes and hemidesmosomes (see Fig. 1.9 ). Individual intermediate filament proteins have characteristic tissue-specific patterns of expression that can be useful for assigning a cell of origin for poorly differentiated tumors. Examples include:
Vimentin , in mesenchymal cells (fibroblasts, endothelium).
Desmin in muscle cells forms the scaffold on which actin and myosin contract.
Neurofilaments are critical for neuronal axon structure and confer both strength and rigidity.
Glial fibrillary acidic protein is expressed in glial cells.
Cytokeratins are expressed in epithelial cells. There are at least 30 distinct different cytokeratins that are expressed in different cell lineages (e.g., lung vs. gastrointestinal epithelium).
Lamins are intermediate filament proteins that form the nuclear lamina, define nuclear shape, and can regulate transcription.
Microtubules are 25-nm-thick fibrils composed of noncovalently polymerized α- and β-tubulin dimers organized into hollow tubes. These fibrils are extremely dynamic and polarized, with “+” and “−” ends. The “−” end is typically embedded in a microtubule organizing center (MTOC or centrosome) near the nucleus, where it is associated with paired centrioles; the “+” end elongates or recedes in response to various stimuli by the addition or subtraction of tubulin dimers. Microtubules:
Serve as mooring lines for molecular motor proteins that use ATP to translocate vesicles, organelles, or other molecules around cells. There are two varieties of these motor proteins, kinesins and dyneins, that typically (but not exclusively) transport cargo in anterograde (− to +) or retrograde (+ to −) directions, respectively.
Mediate sister chromatid segregation during mitosis.
Form the core of primary cilia, single nonmotile projections on many nucleated cells that contribute to the regulation of cellular proliferation and differentiation (mutations in the proteins of the primary cilia complex lead to forms of polycystic kidney disease; see Chapter 20 ).
Can be adapted to form the core of motile cilia (e.g., in bronchial epithelium) or flagella (in sperm).
Cells connect and communicate with each other via junctional complexes that form mechanical links and facilitate receptor-ligand interactions. Similar complexes also mediate interaction with the ECM. Cell-cell junctions are organized into three basic types (see Fig. 1.9 ):
Occluding junctions (tight junctions) seal adjacent epithelial cells together to create a continuous barrier that restricts the paracellular (between cells) movement of ions and other molecules. Occluding junctions form a tight meshlike network (when viewed en face by freeze-fracture electron microscopy) of macromolecular contacts between neighboring cells; the complexes that mediate the cell-cell interactions are composed of transmembrane proteins including the tetraspanning claudin and tight junction–associated MARVEL protein (TAMP) families. These connect to a host of intracellular adaptor and scaffolding proteins, including the three members of the zonula occludens protein family (ZO-1, ZO-2, ZO-3) and cingulin. Besides forming a selectively permeable barrier that seals the space between cells (i.e., the paracellular space), this zone also represents the boundary that separates apical and basolateral membrane domains and helps to maintain cellular polarity. Nevertheless, these junctions are dynamic structures that can be modified to facilitate epithelial healing and inflammatory cell migration across epithelial-lined mucosal surfaces.
Anchoring junctions (adherens junctions and desmosomes) mechanically attach cells—and their cytoskeletons—to other cells or the ECM. Adherens junctions are often closely associated with and beneath tight junctions. Desmosomes are more basal and form several types of junctions. When desmosomes attach the cell to the extracellular matrix (ECM) they are referred to as hemidesmosomes (half a desmosome), since the other half of the desmosome is not present within the ECM. Both adherens junctions and desmosomes are formed by homotypic extracellular interactions between transmembrane glycoproteins called cadherins, on adjacent cells.
In adherens junctions the transmembrane adhesion molecules are associated with intracellular actin microfilaments through which they can also influence cell shape and/or motility. Loss of the epithelial adherens junction protein E-cadherin explains the discohesive invasion pattern of some gastric cancers and lobular carcinomas of the breast ( Chapters 17 and 23 ).
In desmosomes the cadherins are linked to intracellular intermediate filaments, allowing extracellular forces to be mechanically communicated (and dissipated) over multiple cells.
In hemidesmosomes the transmembrane connector proteins are called integrins; like desmosomal cadherins, these attach to intermediate filaments and link the cytoskeleton to the ECM. Focal adhesion complexes are composed of >100 proteins and localize at hemidesmosomes. Their component proteins can generate intracellular signals when cells are subjected to shear stress (e.g., endothelium in the bloodstream or cardiac myocytes in a failing heart).
Communicating junctions (gap junctions) permit the diffusion of chemical or electrical signals from one cell to another. The junction consists of a dense planar array of 1.5- to 2-nm pores (called connexons ) formed by a pair of hexamers (one on each cell) of transmembrane connexin proteins. These form pores that permit passage of ions, nucleotides, sugars, amino acids, vitamins, and other small molecules; permeability of the junction is rapidly reduced by lowered intracellular pH or increased intracellular calcium. Gap junctions play a critical role in cell-cell communication. For example, gap junctions in cardiac myocytes allow cell-to-cell calcium fluxes that allow the many cells of the myocardium to behave as a functional syncytium with coordinated waves of contraction.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here