Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
One of the prevailing models of cancer development proposes that a cancer is initiated and maintained through the function of CSCs, which represent a rare population of cells within a cancer that have an indefinite proliferative potential and are ultimately responsible for the generation of the bulk of cancer cells. This so-called CSC hypothesis has been best studied in hematopoietic malignancies. The ability to purify hematopoietic cells more easily than cells from other tissues, combined with the well-defined cell-surface markers of hematopoietic cells, has allowed the prospective isolation of nearly every hematopoietic cell subset from humans as well as mice.
The CSC hypothesis proposes that cancers are organized into hierarchical populations like normal tissues. At the apex of hierarchy are largely quiescent long-lived CSCs with marked self-renewal capacity that sustain the disease and give rise to the majority of the bulk cancer cells that constitute the disease. While identification of a single normal hematopoietic cell subset as the target of the malignant transformation and the cellular reservoir for disease has been possible for a variety of myeloid leukemias and lymphomas, pinpointing a single cell as the target of malignant transformation has not yet proven possible for other hematopoietic malignancies. In this chapter, we discuss efforts to identify the malignant stem cell for each of the common forms of myeloid and lymphoid leukemias.
The first evidence of a stem cell origin of malignancy came from studies in 1997 performed by Blair et al., as well as Bonnet and Dick in acute myeloid leukemia (AML). These studies demonstrated that most leukemia cells were unable to proliferate extensively and that only a subset of cells was consistently clonogenic. In these studies, a small subset of human Thy1 − CD34 + CD38 − AML cells (0.2% to 1.0%) was identified and shown to be the only cells capable of transferring human AML to immunodeficient mice. In humans, normal hematopoietic stem cells (HSCs) reside in the lineage-negative (Lin − ) CD34 + CD38 − CD90 + CD45RA − compartment and generate multipotent progenitors with lymphomyeloid potential (LMPPs) defined as Lin − CD34 + CD38 − CD90 − CD45RA − cells, as well as more committed myeloid progenitors that are present in the CD34 + CD38 + compartment (see Chapter 9 ). Among the myeloid progenitors, the common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) can be discriminated based on the differential expressions of CD123 (IL3RA), CD110 (MPL), and CD45RA.
The initial observation that AML leukemia-initiating cells (LICs) reside within the CD34 + CD38 − compartment suggested that AML HSCs are rare cells that most closely resemble normal HSCs, sharing a common limited immunophenotype and being a rare population ( Fig. 13.1 ). However, subsequent data have suggested that this conclusion is an oversimplification and that the cell of origin of any myeloid malignancy is likely dictated by a combination of the specific genetic and epigenetic alterations present in the individual patient as well as the cells in which these alterations occur. For example, the aforementioned earliest studies of LICs in AML relied on their transplantation into immunodeficient nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (see box on Evolution of Immunodeficient Mouse Models ) to assay the ability of a defined population of AML cells to give rise to AML in vivo. However, using more immunodeficient xenotransplant models, primary human cells from both CD34 + CD38 − and CD34 + CD38 + compartments have been shown to have LIC activity. In addition, work by Vyas and colleagues has revealed that two expanded populations, both with LIC activity, exist in CD34 + AML (see Fig. 13.1 ). One population shares the immunophenotype of normal LMPPs and the other mirrors the GMP population. The LMPP-like leukemic stem cell (LSC) population can give rise to the GMP-like LSC population, but either can give rise to AML in immunocompromised mice in vivo.
Xenotransplantation of human hematopoietic cells into immunocompromised mice has been an important assay for studying normal and malignant human hematopoiesis for more than 50 years. The first breakthrough in xenograft modeling of human hematopoiesis was the discovery of the severe combined immune-deficient (SCID) mouse. Transplantation of human bone marrow cells into the SCID mouse not only regenerated T and B cells, which the mice lacked genetically, but also continued to generate myeloid progenitors long term. SCID mice are limited by high levels of innate immune function that impede human engraftment, however. To circumvent this limitation, SCID mice were back-crossed onto nonobese diabetic (NOD) mice harboring innate immune defects. NOD-SCID mice support higher levels of human engraftment but are limited by the development of thymic lymphomas and the persistence of functioning natural killer (NK) cells. Both of these problems were addressed by deletion of the IL-2 receptor common gamma chain. Resultant NOD/SCID/gamma-null (NSG) mice have complete loss of B, T, and NK cells; and their defective cytokine signaling also prevents lymphomagenesis. NSG mice support higher levels of human donor cell engraftment and serve as assays that functionally assess the ability of potential cancer stem cells to initiate a malignancy. Newer types of humanized mice, such as the MISTRG mouse, have been developed that express human cytokines that are not cross-reactive with murine cells and are able to stimulate the growth of more human cell types in vivo. Other humanization strategies include constitutive expression of HLA class I and II that strengthens T-cell function or constitutive expression of Sirpa, a transmembrane protein on macrophages that induces tolerance to human grafts. Loss-of-function mutations in KIT proto-oncogene receptor tyrosine kinase (KIT) has allowed for the creation of hematopoietic stem cell (HSC) xenografts without the need for irradiation of the recipient mouse.
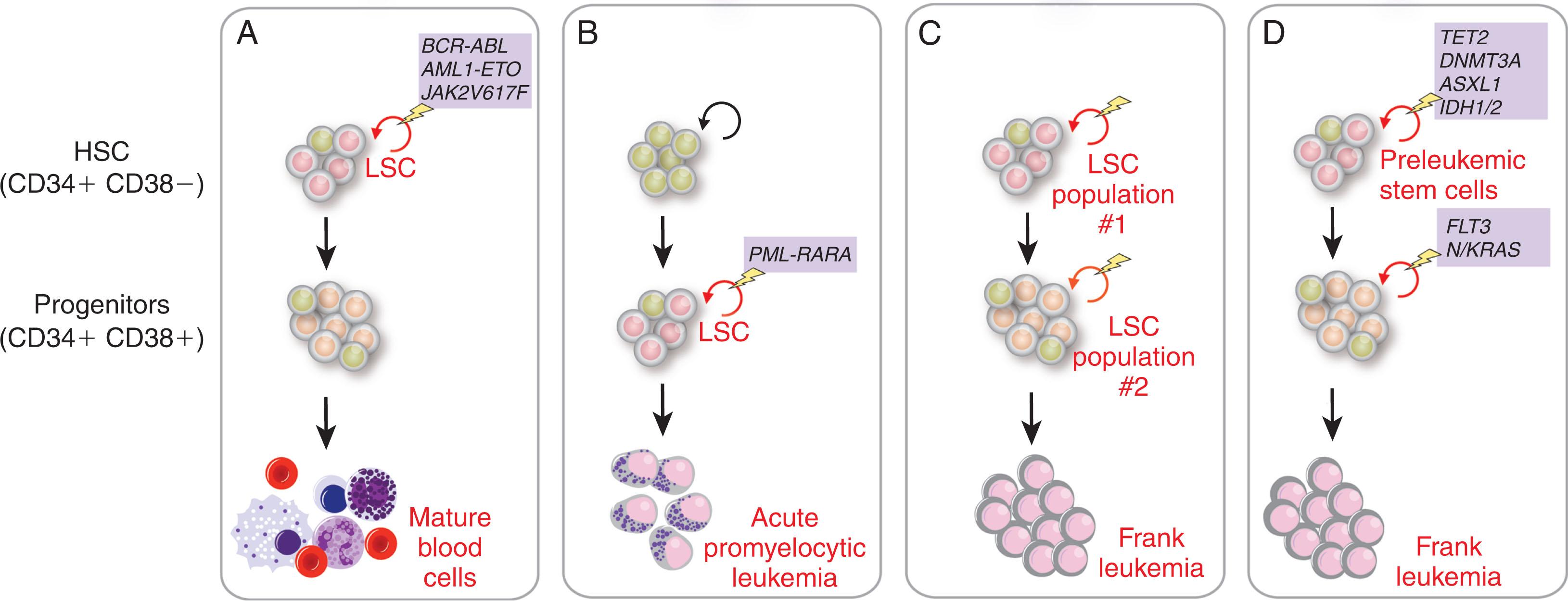
As described in the box on Functional Evaluation of Cell-of-Origin In Vivo , the leukemogenic effects of specific oncogenes directly depend on the specific oncogene as well as the target cell of expression. Based on these facts, consistent LICs may be most easily defined for specific genetically defined subsets of leukemias (such as specific chronic leukemias defined by specific translocations or point mutations) but are much more difficult to define for normal karyotype AML. For example, expression of the AML1-ETO fusion transcript, generated by the common t(8;21) translocation in AML, can be detected not only in leukemic cells but also in normal HSCs from patients in clinical remission from AML. However, these AML1-ETO- expressing HSCs are not leukemic and can differentiate into myeloid and erythroid cells in vitro in a manner similar to HSCs without the AML1-ETO fusion transcripts (see Fig. 13.1 ). Similarly, analysis of mice expressing the AML1-ETO fusion from the endogenous Aml1 locus in vivo has revealed that AML1-ETO -expressing HSCs have aberrant self-renewal capacity but do not develop overt leukemia unless additional genetic abnormalities are present. These data strongly suggest that the acquisition of additional genetic abnormalities in a subset of HSCs or their progeny is required to give rise to overt leukemia. In these studies, the HSCs bearing the AML1-ETO fusion reside within the Lin − CD34 + CD38 − subpopulation that is also the immunophenotype of normal human HSCs, suggesting that the initiating lesion must occur in a cell with an immunophenotype of normal HSCs. However, leukemic cells from 30% to 40% of patients with AML do not express CD34, and LICs from some patients with AML can actually be CD34 − . Interestingly, prior work evaluating the location of the PML-RARA transcript present in acute promyelocytic leukemia (APL) revealed that the PML-RARA translocation is actually present in CD34 − CD38 + populations and not in CD34 + CD38 − HSC-enriched populations (see Fig. 13.1 ). These data clearly reveal that there is enormous heterogeneity in the cell-of-origin of AML.
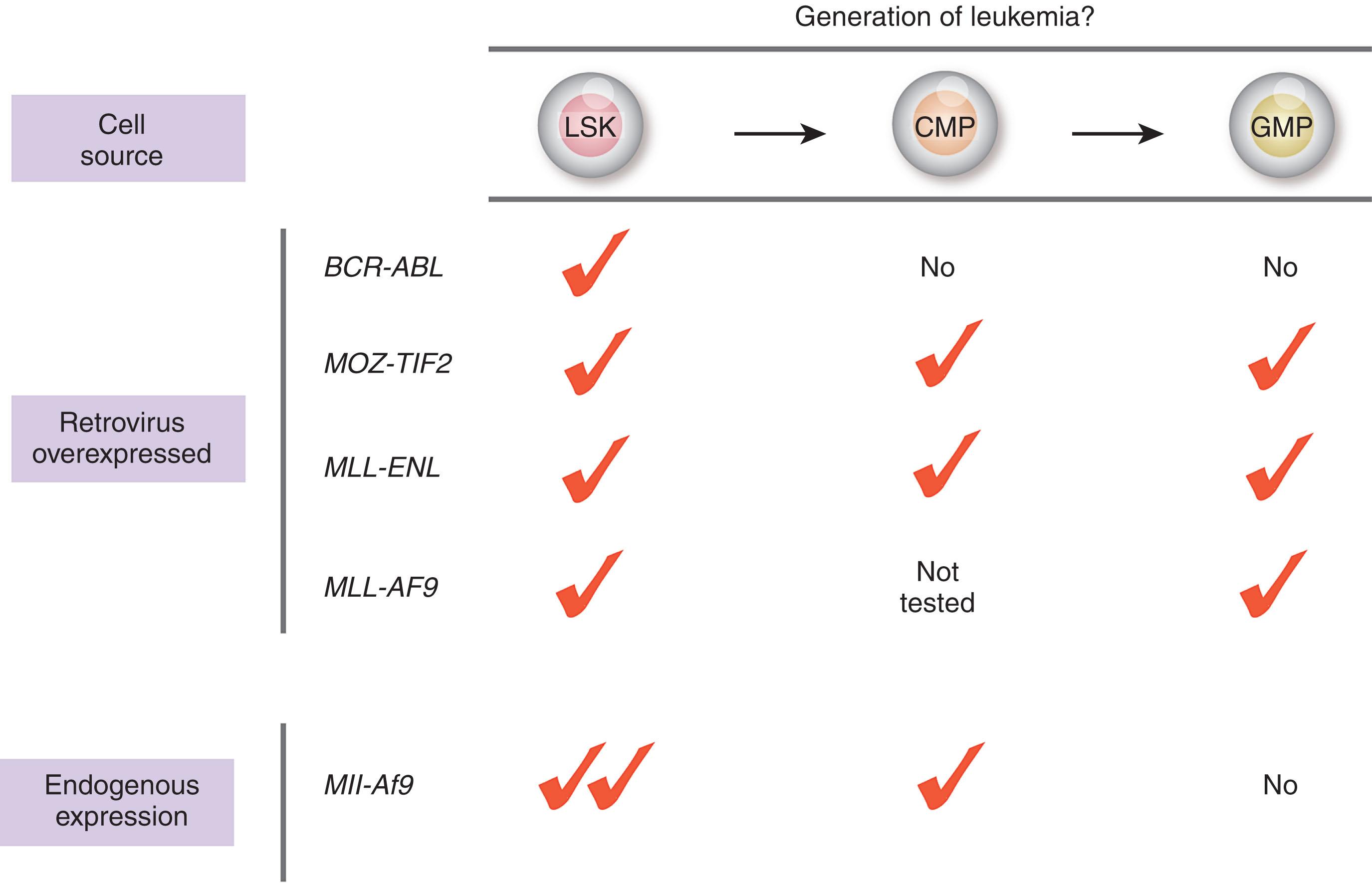
The earliest studies of cancer stem cells in acute myeloid leukemia (AML) suggested that AML is initiated by genetic alterations that take place in hematopoietic stem cells (HSCs). Alternatively, however, there is clear evidence that some leukemias may be initiated by mutations arising in more committed progenitors that provide these cells with aberrant self-renewal capacity that they otherwise lack. Numerous studies have attempted to directly address each of these possibilities through selective expression of specific oncogenes in isolated HSCs, common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and even more mature myeloid cells ( Fig. 13.2 ). Such experiments have been most thoroughly performed using the expression of mixed lineage leukemia (MLL) fusion oncoproteins. MLL encodes an epigenetic enzyme required for normal hematopoiesis due to its role in the maintenance of HOX gene expression (see Chapter 9 ). Translocation events fusing the N-terminus of MLL to over 50 different C-terminal partners are common in both AML and acute lymphoblastic leukemia (ALL). Overexpression of the MLL-ENL fusion oncoprotein in self-renewing HSCs as well as in myeloid progenitor populations with more restricted self-renewal potential including CMPs and GMPs result in the rapid onset of AML in vivo. Similar results have been seen with retroviral overexpression of a different MLL fusion oncoprotein, MLL-AF9 , as well as an unrelated AML-associated fusion, MOZ-TIF2 (see Fig. 13.2 ). In contrast, overexpression of BCR-ABL is only able to transform HSCs but not committed downstream progenitors. These results unequivocally demonstrate (1) the ability of an MLL fusion oncoprotein to convert a myeloid progenitor cell population that is distinct from an HSC to acquire leukemogenic self-renewal activity and (2) that the cell-of-origin where a genetic alteration is expressed may regulate the resultant ability for a malignant disease to develop. It is also important to note that when the MLL-AF9 fusion is expressed under endogenous regulatory control, Mll-Af9 transforms HSCs but not GMPs. These data clearly reveal that the dosage of oncogene expression may also regulate cellular transformation.
Advancement in techniques to map genetic alterations in cancer has allowed for much finer tracking of somatic mutations in AML and other hematopoietic malignancies with a normal karyotype. It is now believed that an average of five coding mutations is present in adults with de novo AML (see Chapter 59 ). Several groups have now studied the occurrence of somatic mutations in bulk AML cells and the remaining seemingly nonaffected HSCs. This work has clearly shown that the HSC compartment in patients with AML contains HSCs with none of the mutations found in the AML as well as HSCs with various combinations of genetic alterations similar to that present in the bulk malignant cells (see Fig. 13.1 ). These latter HSCs are now understood to be “preleukemic stem cells” that initiate AML and can also be identified in remission samples, indicating that they are able to survive induction chemotherapy (see box on Preleukemic Stem Cells). Many of the mutations that occur in preleukemic HSCs confer growth properties that allow them to outcompete normal HSCs and presumably lead to relapse. Interestingly, mutations occurring in preleukemic HSCs are enriched in genes regulating DNA methylation, chromatin modifications, and the cohesin complex, while genetic alterations activating signaling are often present in more downstream overt malignant cells and absent from preleukemic HSCs. A series of recent studies performing single-cell mutational analysis of AML have further solidified these findings and identified that mutations in signaling genes (such as FLT3, NRAS, and KRAS) are often acquired in subclones of AML and can be acquired in multiple distinct subclones.
Hematopoietic stem cells (HSCs) are rare (~1–2 per 10 8 bone marrow cells) and quiescent cells that rarely divide. However, all blood cells originate from progenitor cells that in turn originate from HSCs and thus HSCs must replicate/self-renew to continue the cycle. Mutations in HSCs can therefore occur during cell division and accumulate, sometimes speeding self-renewal divisions and leading to clonal hematopoiesis. Although able to confer a growth advantage to clones derived from mutated HSCs, these single mutations alone are not sufficient to transform cells, resulting in an overt malignant phenotype. Because growth-promoting gene mutations are common in cancer, it is thought that HSCs with mutations that cause clonal hematopoiesis might exist as preleukemic stem cells in patients with leukemia for months to years prior to diagnosis (see Chapter 59 ). Cooperative driver mutations are then acquired that cause cancer to develop. Acquired somatic mutations causing clonal hematopoiesis exist in healthy individuals and increase in frequency with age. The risk of hematologic malignancy is higher in patients with clonal hematopoiesis compared with matched controls, providing evidence for leukemic predisposition (see Chapter 59 ). The most common mutations causing clonal hematopoiesis were found in the genes for DNMT3A , TET2 , SRSF2, SF3B1, U2AF1, IDH1/2 , and ASXL1 which are also known to be recurrently mutated in myeloid malignancies. These genes encode proteins involved in epigenetic modifications of chromatin and DNA as well as in RNA splicing. Functional experiments have shown that DNMT3A and TET2 actually increase the number of HSCs in the bone marrow due to impaired differentiation and increased self-renewal, respectively. Studies of a few patients with preleukemic stem cells who went on to develop leukemia have shown that at least in some cases, the leukemia developed only after acquisition of another driver mutation such as mutations in FLT3 . Additionally, different somatic mutations drive distinct patterns of clonal hematopoiesis, such as mutations in the splicing factors U2AF1, SF3B1, and SRSF2, which exclusively occur in patients over the age of 70 and may only confer a growth advantage under the selection pressures of an aging hematopoietic system.
Similar to AML, tracking major chromosomal abnormalities allowed earlier investigators to establish that clonal hematopoiesis in MDS originated in HSCs (see Chapter 61 ). Initially, deletion of the long arm of chromosome 5 (del(5q)) was identified in the HSCs of patients with del(5q)-MDS. Moreover, del(5q) CD34 + CD38 − cells were shown to possess MDS-initiating potential using in vitro and in vivo stem cell assays. Systems subsequently improved phenotypic identification of HSCs that have allowed investigators to focus on Lin − CD34 + CD38 − CD90 + CD45RA − cells as candidate CSCs in MDS. These studies showed that in patients with del(5q)-MDS, 99% of Lin − CD34 + CD38 − CD90 + CD45RA − cells contained del(5q) and that they were molecularly and functionally distinct from clonally involved GMPs and MEPs. By gene expression profiling and principle component analysis of RNA sequencing data, the three cell populations were shown to be distinctively different, with the Lin − CD34 + CD38 − CD90 + CD45RA − cells selectively expressing genes characteristic of normal HSCs. These candidate stem cells were exclusively able to sustain long-term generation of MDS myeloid progenitors in vitro that was not observed with GMPs, MEPs, or CMPs. Moreover, patients with del(5q)-MDS were found to have residual del(5q) clones in the CD34 + CD38 − CD90 + stem cell compartment during clinical remission. Over time, most of these patients experienced expansion of the clone, leading to cytogenetic and clinical progression.
The evidence for an MDS-initiating stem cell is not limited to isolated del(5q)-MDS, which may be a unique disease entity given its distinct clinical features. In fact, recent studies have shown that recurrent driver mutations occur at the HSC level in a broad panel of low to intermediate risk MDS and that the 5q deletion preceded any other identifiable recurrent driver mutations in isolated del(5q)-MDS. The same was observed with SF3B1 mutation-positive MDS that is characterized by ringed sideroblasts, where SF3B1 mutations are thought to be early and potentially initiating events. Targeted screening for mutations of a large number of genes known to be frequently mutated in MDS and other myeloid malignancies has been performed on bulk bone marrow from patients with MDS. After identifying the mutations specific to each patient, the different cell compartments (HSC, GMP, or MEP) were purified and the screen for mutations was repeated. It was hypothesized that if progenitor cells acquired self-renewal capacity, allowing them to persist long enough for the mutations to be responsible for the MDS phenotype, then the mutations would be identifiable in progenitors but not within HSCs. However, all of the mutations in MDS patients have been traced back to the HSC compartment (Lin − CD34 + CD38 − CD90 + CD45RA − cells) and all of the mutations found in the Lin − CD34 + CD38 − CD90 + CD45RA − MDS-initiating cells have also been identified in downstream GMPs and MEPs. These findings suggest that MDS is propagated by a stem cell that develops and acquires mutations at an early stage in the disease.
Interestingly, Chen and colleagues provided evidence for a non-linear, parallel manner by which MDS patient HSCs progress to AML. They identified distinct progenitor populations that gave rise to MDS and MDS-related AML. They demonstrated that the dominant AML clone originated from a clone that was included in as pre MDS and/ or MDS HSCs but was undetectable in the MDS blast cells, indicating MDs stem cells are distinct from those that contribute to AML that follow MDS.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here