Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
High-throughput array technologies such as genomics and proteomics have provided unprecedented ability to personalize output of biomarkers implicated in select disease states.
Pharmacogenomics, through utilization of novel technologies, can detect unique genomic alterations to serve as diagnostic, prognostic, or predictive biomarkers, which relate to an individual or group response to particular therapies.
Elucidation of unique genomic markers related to personal responses can tailor therapy to the individual, thereby maximizing efficacy and minimizing side effects.
Interpretation of pharmacogenomics data can be discretionary in that many genotypic and phenotypic variables are often involved.
Pharmacogenomics is the study of how genetic variations influence individual response to drugs, in terms of efficacy and risk for adverse drug reactions. This relatively new field combines pharmacology (the study of drugs and their uses, effects, and mode of action) and genomics (the study of genes and their functions) to develop effective, safe medications and doses that are tailored to a person’s genetic makeup. Genetic variations influencing individual response to drugs can be generally grouped into germline genetic polymorphisms and somatic mutations as occur in tumor tissues. Germline genetic polymorphisms in genes encoding drug-metabolizing enzymes, transporters, drug targets, and human leukocyte antigens (HLAs) have been associated with interindividual differences in the pharmacokinetics, efficacy, and adverse drug reactions of many drugs. Somatic mutations in tumor tissues are important determinants of the tumor sensitivity or resistance to many molecular targeted agents. The knowledge of how inherited differences in genes affect the body’s response to medications will allow the development of personalized medicine that will provide greater efficacy and safety in drug development and therapy. This chapter summarizes recent important findings on pharmacogenomics of drug-metabolizing enzymes, transporters, drug targets (including germline genetic polymorphisms and somatic mutations), and HLAs, and discusses their implications in the development of personalized medicine to treat a wide range of health problems.
The concept of a relationship between a substance and its effect on genes, rudimentary pharmacogenetics, has been present since the early 1900s. Dr. Archibald Garrod described that genetic variations could cause adverse biological reactions when chemical substances were ingested ( ). Garrod’s work on alcaptonuria constituted the first proof of mendelian genetics in humans. As a result of these studies, he advanced the hypothesis that genetically determined differences in biochemical processes could affect outcome after drug administration ( ). The idea of pharmacogenomics, also referred to as toxogenomics, is considered to have matured from pharmacogenetics, a discipline comprising genetics, pharmacology, and biochemistry, as a fusion of pharmacogenetics and genomics. Whereas pharmacogenetics classically investigates the hereditary impacts upon the action of drugs, pharmacogenomics extends pharmacogenetics to include the multifactorial drug responses caused by many genetic alterations plus environmental factors ( ). The recognition of these complex interactions and the advance of genetics into genomics via high-throughput analysis of gene interrogation algorithms have given rise to the application of this science as a means of personalizing drug effects to a group or individual. The term personalized medicine has been coined toward the promise of making therapy safer and more effective, and by giving drugs that fit a person’s genes.
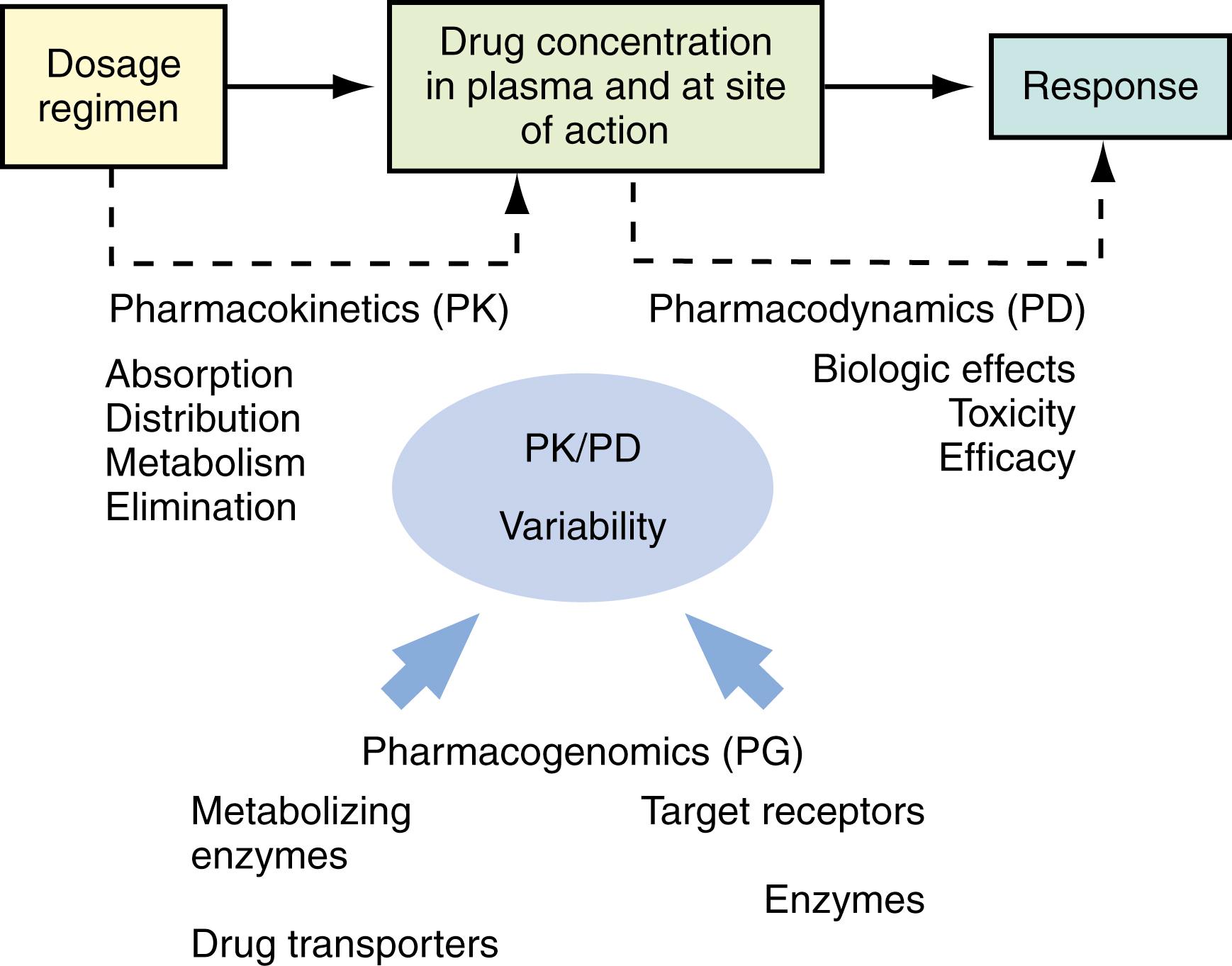
The events following a drug administration can be divided into two phases: pharmacokinetics and pharmacodynamics . Pharmacokinetics relates a dosage regimen to drug concentrations achieved with time; pharmacokinetics concerns drug absorption, distribution, metabolism, and excretion. Pharmacodynamics relates drug concentrations to the magnitude of the desired or adverse effects produced with time; pharmacodynamics concerns drug target interaction, downstream signaling events, and pharmacologic response. Each drug, after it enters the body, interacts with numerous proteins, including carrier proteins, transporters, metabolizing enzymes, and target receptors. These protein interactions determine the pharmacokinetics (i.e., absorption, distribution, metabolism, and excretion) and pharmacodynamics (i.e., target site of action and pharmacologic effects) of a drug. As a result, the overall response to a drug is determined by the interplay of multiple genes that are involved in the pharmacokinetic and pharmacodynamic pathways of a drug ( Fig. 75.1 ). The topics that follow summarize recent findings with regard to the functional and clinical relevance of genetic variations of drug-metabolizing enzymes, transporters, drug targets, and HLAs.
Drug-metabolizing enzymes are proteins that catalyze the biochemical modifications of xenobiotics (e.g., drugs) and endogenous chemicals (e.g., hormones, neurotransmitters). Drug metabolism can result in the activation (toxication) or deactivation (detoxication) of the chemical. Even though both occur, the majority of metabolites of most drugs are deactivation products. Broadly, drug-metabolizing enzymes are divided into two categories: phase I (functionalizing) enzymes that introduce or remove functional groups in a substrate through oxidation, reduction, or hydrolysis; and phase II (conjugating) enzymes that transfer moieties from a cofactor to a substrate. Essentially all of the major human metabolizing enzymes exhibit genetic polymorphisms at the genomic level, and many of these enzymes have clinically relevant genetic polymorphisms ( Fig. 75.2 ). A gene is considered to be polymorphic when the frequency of a variant allele in a specific population is at least 1%.
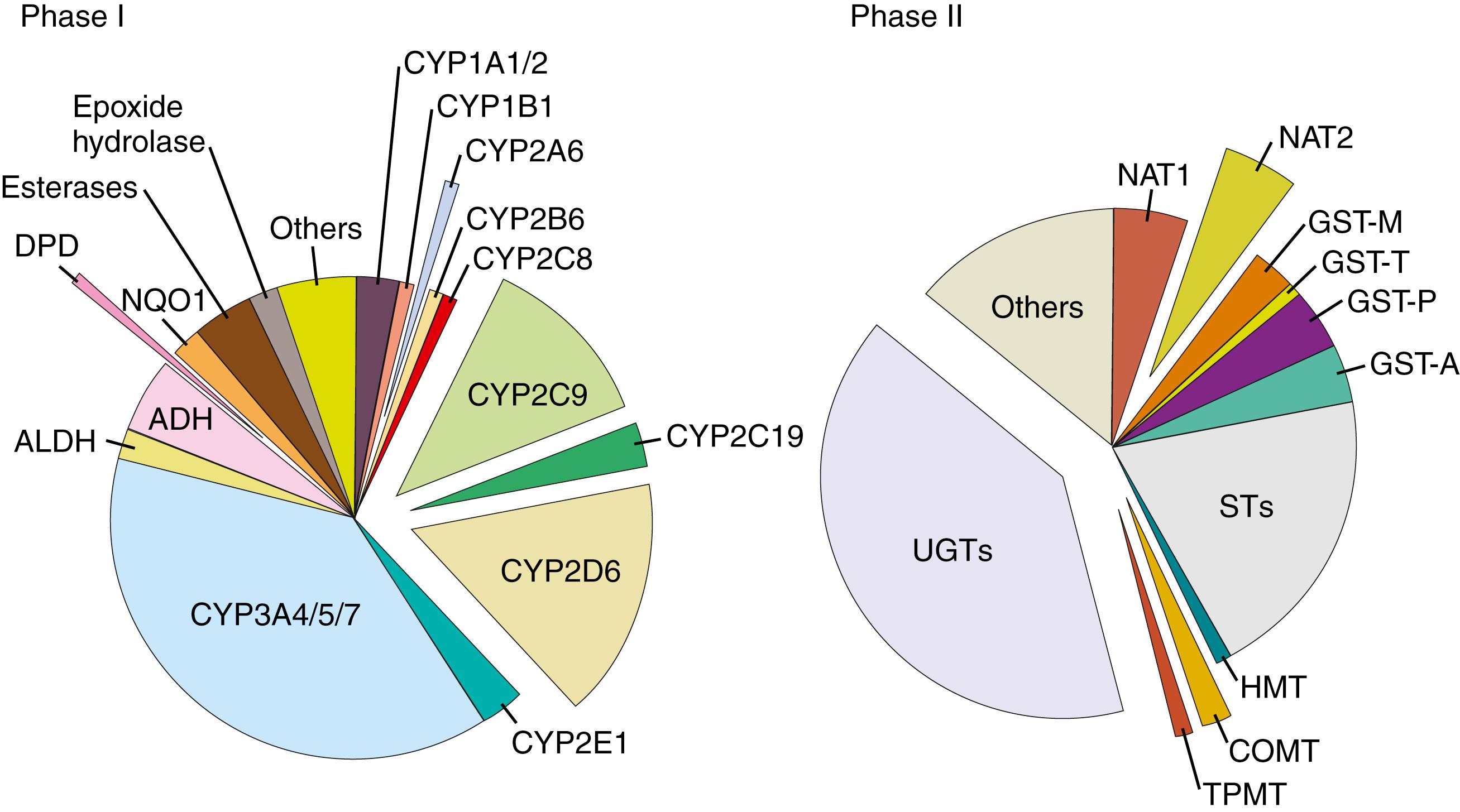
Phase I metabolizing enzymes include those involved in oxidation (e.g., cytochrome P450 [CYP], alcohol dehydrogenase, aldehyde dehydrogenase, dihydropyrimidine dehydrogenase, monoamine oxidase, and flavin-containing monooxygenase), reduction (e.g., nicotinamide adenine dinucleotide phosphate [NADPH]-cytochrome P450 reductase and reduced cytochrome P450), and hydrolysis (e.g., epoxide hydrolase, esterases, and amidases).
The most important phase I enzymes that exhibit clinically relevant genetic polymorphisms are in the CYP superfamily, which represents the most important system responsible for catalyzing the oxidation of a large number of endogenous and exogenous compounds, including drugs, toxins, and carcinogens. In this superfamily, 57 genes and 58 pseudogenes have been identified, which are divided into 18 families and 43 subfamilies. Among them, three CYP subfamilies—CYP1, CYP2, and CYP3—are the main ones contributing to the oxidative metabolism of more than 90% of clinically used drugs.
The human CYP genes are highly polymorphic. The polymorphisms within the CYP genes, which include gene deletions, missense mutations, deleterious mutations creating splicing defects or premature stop codon, and gene duplications, can result in abolished, reduced, normal, or enhanced enzyme activity. Based on the level of CYP enzyme activity, patients can be classified into four phenotypes: poor metabolizer (abolished activity), intermediate metabolizer (reduced activity), extensive metabolizer (normal activity), and ultrarapid metabolizer (enhanced activity). Poor metabolizers may have higher concentrations of a drug that is inactivated by that enzyme pathway, thereby requiring a lower dose to avoid adverse reactions; whereas ultrarapid metabolizers may require a higher dose to achieve effective therapeutic drug concentrations. The opposite pattern of reactions occurs in response to a prodrug that requires metabolic activation to exert its pharmacologic effect. A prodrug may have little therapeutic effect in poor metabolizers while producing toxic levels of active form in ultrarapid metabolizers.
It is evident that genetic polymorphisms within the CYP genes have significant impact on drug disposition and/or response. The common functional polymorphisms in the major human CYP genes and their clinical relevance are summarized in Table 75.1 . Comprehensive reviews on the pharmacogenetics of CYPs have been published ( ; ; ; ; ). Notably, the most pharmacologically and clinically relevant polymorphisms are found in CYP2D6, CYP2C9, and CYP2C19 . Of the US Food and Drug Administration (FDA)–approved drug labels referring to human genomic biomarkers, 62% pertain to polymorphisms in the CYP enzymes, with CYP2D6 (35%), CYP2C19 (17%), and CYP2C9 (7%) being the most common ( ). For a review, see .
| Common Allelic Variants | Polymorphism/Substitution | ALLELE FREQUENCY (%) a | Functional Effect b | Highlights of Clinical Relevance c | ||
|---|---|---|---|---|---|---|
| Ca | As | Af | ||||
| CYP1A1 | Mainly expressed in extrahepatic tissues. | |||||
| CYP1A1∗2A | 3698T>C(MspI) | 6.6-19 | 33-54 | 22-28 | ↑Inducibility | CYP1A1, 1A2, and 1B1 play an important role in the bioactivation of a variety of carcinogens. |
| CYP1A1∗2B | I462V; 3698T>C(MspI) | – | – | – | ↑Inducibility | ↑Lung cancer risk generally associated with highly inducible or active CYP1A1 polymorphisms such as CYP1A1∗2C. |
| CYP1A1∗2C | I462V | 2.2-8.9 | 28-31 | 0-2.7 | ↑Activity | CYP1A1 genotypes also associated with risk for breast, prostate, and ovarian cancers that are possibly related to estrogen activation. |
| CYP1A1∗3 | 3204T>C | 0 | 0 | 7.6-14 | Normal | |
| CYP1A1∗4 | T461N | 2.0-5.7 | – | – | Normal | |
| CYP1A2 | CYP1A2 accounts for ∼13% of total hepatic CYP content. | |||||
| CYP1A2∗1C | –3860G>A | ↓Inducibility | High inducible ∗ 1F genotype associated with ↑clearance of CYP1A2 substrates (e.g., caffeine) after smoking or omeprazole treatment. | |||
| CYP1A2∗1F | –163C>A | 33 | 68 | ↑Inducibility | ∗ 1K associated with ↓in vivo caffeine metabolism. | |
| CYP1A2∗1K | Haplotype (–63C>A, –739T>G, –729C>T) | 0.5 | ↓Inducibility ↓Activity |
CYP1A2 genotypes are associated with cancer risk. | ||
| CYP2A6 | CYP2A6 accounts for 1% to 10% of total hepatic CYPs. | |||||
| CYP2A6∗1X2 | Gene duplication | 1.7 | 0.4 <1 |
↑Activity ↓Activity |
The frequency of CYP2A6 alleles has marked ethnic differences. CYP2A6∗4 accounts for the majority of PMs in Asians. | |
| CYP2A6∗2 CYP2A6∗4 |
L160H Gene deletion |
1-3 0.5-1 |
7-22 | 15-20 | Abolished activity | Because nicotine is converted to cotinine by CYP2A6, a high expression/activity of CYP2A6 is proposed to increase the susceptibility to nicotine addiction and the risk for tobacco-related cancers. Therefore CYP2A6 genetic variation could play a role in nicotine addiction and tobacco-related cancer risks. |
| CYP2B6 | CYP2B6 is mainly expressed in the liver, accounting for 6% of total CYPs. | |||||
| CYP2B6∗4 | K262R | 5 | ↑Activity | The anticancer drug cyclophosphamide (CPA) is bioactivated by CYP2B6. CYP2B6 polymorphisms would likely affect the PK and/or PD of CPA. For example, CYP2B6∗6 carriers exhibited ↑CPA clearance and CPA 4-hydroxylation activity. | ||
| CYP2B6∗5 CYP2B6∗6 CYP2B6∗7 |
R487C Q172H; K262R Q172H; K262R; R487C |
11-14 16-26 13 |
1 16 0 |
↓Expression ↑Activity ↑Activity |
CYP2B6 polymorphisms may affect the PK and therapeutic outcome of anti-HIV agents such as efavirenz and nevirapine. For example, the CYP2B6 Q172H variant is associated with ↑plasma concentrations of efavirenz and nevirapine. | |
| CYP2C8 | CYP2C8 accounts for ∼7% of total hepatic CYP contents. | |||||
| CYP2C8∗2 CYP2C8∗3 CYP2C8∗4 |
I269F R139K; K399R I264M |
0.4 13 7.5 |
0 | 18 2 |
↓Activity ↓Activity ↓Activity |
CYP2C8∗3 is associated with ↓clearance of both R- and S-ibuprofen. |
| CYP2C9 | CYP2C9 accounts for ∼20% of total hepatic CYP contents. | |||||
| CYP2C9∗2 CYP2C9∗3 CYP2C9∗5 |
R144C I359L D360E |
13-22 3-16 0 |
0 3 2 |
3 1.3 0 |
↓Activity ↓Activity ↓Activity |
CYP2C9∗2 and ∗3 have been shown to affect the oral clearance of at least 17 different CYP2C9 substrate drugs (e.g., S-warfarin, celecoxib, ibuprofen, and phenytoin). |
| CYP2C19 | The PM phenotype of CYP2C19 occurs in 12% to 23% of the Asian population, 1% to 6% of Caucasians, and 1% to 7.5% of black Africans. | |||||
| CYP2C19∗2 CYP2C19∗3 CYP2C19∗17 |
Splicing defect; I331V W212X; I331V I331V |
15 0.04 18 |
30 5 4 |
17 0.4 |
Abolished activity Abolished activity ↑Transcription |
Polymorphisms in the CYP2C19 gene are known to affect the PK and/or response of several classes of drugs, including proton pump inhibitors (e.g., omeprazole) and barbiturates. |
| CYP2D6 | CYP2D6 accounts for ∼2% of total hepatic CYP contents. However, it is involved in the metabolism of ∼25% of all drugs in clinical use. | |||||
| CYP2D6∗3 | Frameshift | 1-2 | <1 | Abolished activity (PM) | Unlike other CYPs, CYP2D6 is not inducible, and thus genetic polymorphisms are largely responsible for the variation in enzyme expression and activity. | |
| CYP2D6∗4 | Splicing defect | 20-25 | 1 | 6-7 | Abolished activity (PM) | CYP2D6 genotypes exhibit large interethnic differences: low frequency of PM in Asian (∼1%) and African (0–5%) populations, compared to Caucasians (5–14%). |
| CYP2D6∗5 | Gene deletion | 4-6 | 4-6 | 4-6 | Abolished activity (PM) | The CYP2D6 genotype is of great importance for the PK and response of many drugs, including tricyclic antidepressants, antiarrhythmics, neuroleptics, analgesics, antiemetics, and anticancer drugs. |
| CYP2D6∗10 | P34S; S486T | <2 | 50 | 3-9 | ↓Activity (IM) | |
| CYP2D6∗17 | T107I; R296C; S486T | <1 | 20-34 | ↓Activity (IM) | ||
| CYP2D6∗41 | R296C; splicing defect; S486T | 1.3 | 2 | 5.8 | ↓Activity (IM) | |
| CYP2D6∗1×N, N≥2 | Gene duplication | ↑Activity (UM) | ||||
| CYP2D6∗2×N, N≥2 | Gene duplication | ↑Activity (UM) | ||||
| CYP3A4 | CYP3A4 has the highest abundance in the human liver (∼40%) and metabolizes over 50% of all currently used drugs. | |||||
| CYP3A4∗1B | 5′ flanking region | 2-9 | 0 | 35-67 | Altered expression | Genetic polymorphisms in CYP3A4 appear to be more prevalent in Caucasians than in Asians. |
| CYP3A4∗2 | S222P | 2.7-4.5 | 0 | 0 | Substrate-dependent altered activity | There is no consensus on a direct functional or clinical association of CYP3A4 polymorphism. CYP3A4 polymorphism may have minor or moderate clinical relevance. |
| CYP3A4∗3 | M445T | 1.1 | ↓Activity | |||
| CYP3A4∗17 | F189S | 2.1 | ↓Activity | |||
| CYP3A4∗18 | L293P | 0 | 1 | ↑Activity | ||
| CYP3A5 | The clinical relevance of the CYP3A5 polymorphism is demonstrated by the fact that the PK of the immunosuppressive drug tacrolimus is associated with the CYP3A5 genotype. | |||||
| CYP3A5∗3 | Splicing defect | 90 | 75 | 50 | Abolished activity | |
| CYP3A5∗6 | Splicing defect | 0 | 0 | 7.5 | Severely ↓activity | |
| CYP3A5∗7 | 346 frameshift | 0 | 0 | 8 | Severely ↓activity | |
| CYP3A7 | CYP3A7 is a predominantly fetal enzyme. | |||||
| CYP3A7∗1C CYP3A7∗2 |
Promoter T409R |
3 8 |
28 | 6 62 |
↑Expression ↑Activity |
The in vivo functional effect of CYP3A7 polymorphism is demonstrated by the fact that carriers of the CYP3A7∗1C allele had a significantly decreased endogenous level of dehydroepiandrosterone sulfate, a specific substrate of CYP3A7. |
a Allele frequency data are obtained from , , , , , , .
b Functional effect data are obtained from the Human Cytochrome P450 (CYP) Allele Nomenclature Committee website ( http://www.cypalleles.ki.se/ ).
Unlike other CYPs, CYP2D6 is not inducible. Therefore variations in the enzyme expression and activity are largely attributable to the genetic polymorphisms. The CYP2D6 gene is highly polymorphic with more than 63 functional variants identified to date ( https://www.pharmvar.org/ ). These alleles result in abolished, decreased, normal, or ultrarapid CYP2D6 enzyme activity. The most important null alleles are CYP2D6∗4 (splicing defect) and CYP2D6∗5 (gene deletion); the common alleles with severely reduced enzyme activity are represented by CYP2D6∗10 , ∗17 , and ∗41 ; ultrarapid enzyme activity is derived from duplication or multiduplications of active CYP2D6 genes (e.g., CYP2D6∗1 × N [N ≥2]) (see Table 75.1 ). The distributions of CYP2D6 alleles exhibit notable interethnic differences. The nonfunctional allele CYP2D6∗4 is prevalent in Caucasians (allelic frequency, ∼25%); the reduced function allele CYP2D6∗10 is common in Asians (allelic frequency, ∼40%); and CYP2D6∗17 is common in Africans (allelic frequency, ∼34%) ( ). As a result, CYP2D6 poor metabolizers, mainly derived from null allele CYP2D6∗4 , have a high frequency in Caucasians (5–14%) compared with Africans (0–5%) and Asians (0–1%). In contrast, CYP2D6 ultrarapid metabolizers due to gene duplication or multiduplications have a high frequency in Saudi Arabians (20%) and black Ethiopians (29%) compared with Caucasians (1–10%) ( ). The interethnic difference in the CYP2D6 genotypes may contribute to the interethnic variation in the disposition and response of substrate drugs. Although it accounts for ∼2% of total hepatic CYP contents, CYP2D6 is involved in the metabolism of ∼25% of all drugs in clinical use. CYP2D6 genotype is of great importance to the pharmacokinetics and response of many drugs, including tricyclic antidepressants, antiarrhythmics, neuroleptics, analgesics, antiemetics, and anticancer drugs (for a review, see ). A representative example is the implication of CYP2D6 phenotypes to the pharmacokinetics and clinical efficacy of tamoxifen, a prodrug requiring metabolic activation primarily mediated by CYP2D6. The pharmacogenetics of tamoxifen is discussed in detail later.
The human CYP2C9 and CYP2C19 genes are highly homologous at the nucleotide level. The most common nonsynonymous CYP2C9 polymorphisms, CYP2C9∗2 and CYP2C9∗3, result in the enzyme with differing affinity or intrinsic clearance for different substrates. CYP2C9∗2 effect appears substrate specific, while CYP2C9∗3 variant generally leads to reduced catalytic activity toward the majority of CYP2C9 substrates. The clinical importance of CYP2C9 polymorphisms is exemplified by the CYP2C9 genotype-guided dosing of an oral anticoagulant warfarin. Patients carrying either CYP2C9∗2 or CYP2C9∗3 require significantly smaller daily dose of warfarin to maintain desired therapeutic effects while avoiding severe toxicity, as compared with patients carrying the wild-type CYP2C9 ( ). For CYP2C19, a splice site mutation in exon 4 (CYP2C19∗2) and a premature stop codon in exon 4 (CYP2C19∗3) represent the two most predominant null alleles. By genotyping for CYP2C19∗2 and ∗3, ∼84%, ∼100%, and >90% of poor CYP2C19 metabolizers could be detected in Caucasians, Asians, and Africans, respectively. CYP2C19 poor metabolizers are found in 12% to 23% of Asians, 1% to 6% of Caucasians, and 1% to 7.5% of Africans. CYP2C19 genotypes are clinically relevant to the pharmacokinetics and response of several classes of drugs, including proton pump inhibitors (e.g., omeprazole) and barbiturates (for a review, see ).
The most important phase II enzymes that exhibit functional and clinically relevant genetic polymorphisms are uridine diphosphate glucuronosyltransferase (UGT), sulfotransferase (SULT), glutathione S-transferase (GST), N -acetyltransferase (NAT), and thiopurine methyltransferase (TPMT) (see Fig. 75.2 ). Table 75.2 summarizes common functional polymorphisms in these phase II enzymes and highlights their clinical significance.
| Allelic Variants | Polymorphism/Substitution | ALLELE FREQUENCY (%) | Functional Effect | Highlights of Clinical Relevance | ||
|---|---|---|---|---|---|---|
| Ca | As | Af | ||||
| UGT1A1 a | UGT1A1 low promoter activity alleles (e.g., UGT1A1∗28) are significantly associated with ↓glucuronidation of SN-38 (the active metabolite of irinotecan), thereby resulting in ↑risk for irinotecan-induced toxicity. | |||||
| UGT1A1∗6 | G71R | 0 | 13-23 | – | ↓Activity | Genetic variations in UGT1A1 may modify susceptibility to steroid-related cancers including breast, ovarian, endometrial, and prostate cancers. |
| UGT1A1∗28 | (TA)6>(TA)7 in promoter | 29-40 | 13-16 | 36-43 | ↓Expression | |
| UGT1A1∗33 | (TA)6>(TA)5 in promoter | 0-0.7 | 0 | 3-8 | ↑Expression | |
| UGT1A1∗34 | (TA)6>(TA)8 in promoter | 0-0.7 | 0 | 0.9-7 | ↓Expression | |
| UGT1A6 a | UGT1A6 catalyzes the glucuronidation of aspirin and acetaminophen. | |||||
| UGT1A6∗2 | T181A, R184S | 30 | 23 | ↓Activity | “Low activity” UGT1A6 variants, leading to increased salicylate levels in aspirin users, are associated with a lower risk for colon cancer. | |
| UGT1A6∗3 | R184S | 1-2 | 1.6 | Unknown | ||
| UGT1A6∗4 | T181A | 2.4 | Unknown | |||
| UGT1A7 a | ||||||
| UGT1A7∗2 | N129K, R131K | 24-34 | 15 | 39 | Similar activity | UGT1A7 is an important extrahepatic UGT that inactivates a variety of carcinogens. |
| UGT1A7∗3 UGT1A7∗4 |
N129K, R131K, W208R W208R |
23-36 1-1.7 |
26 0 |
23 1 |
↓Activity ↓Activity |
Low-activity UGT1A7 variants increase the risk of developing tobacco-related cancers, specifically orolaryngeal cancer. |
| UGT2B7 a | UGT2B7 is of major significance for the glucuronidation of a number of clinically important drugs (e.g., morphinan derivatives, epirubicin, and zidovudine). | |||||
| UGT2B7∗2 | H268Y | 49-54 | 27 | Similar or decreased activity | Further studies are needed to elucidate the clinical impact of the UGT2B7 polymorphism. | |
| UGT2B15 a | UGT2B15 is the most efficient UGT2B involved in the inactivation of steroid hormones, mainly androgens. | |||||
| UGT2B15∗2 | D85Y | 52-55 | 36-49 | 39 | ↑Activity | UGT2B15 polymorphisms have a potential role in a modified risk for prostate cancer. |
| SULT1A1 b | SULT1A1 is the most highly expressed hepatic SULT. | |||||
| SULT1A1∗2 | R213H | 25-36 | 4.5-17 | 27-29 | ↓Activity and ↓thermal stability | SULT1A1 plays an important role in the sulfation of the metabolites of tamoxifen, 4-hydroxy-tamoxifen, and endoxifen. SULT1A1∗2 is associated with decreased survival of breast cancer patients treated with tamoxifen. |
| SULT1A1∗3 | M223V | 1.2 | 0.6 | 23 | Similar activity | |
| GST c | ||||||
| GSTA1∗B | Promoter point mutation (T-631G, T-567G, C-69T, G-52A) | 40 | 41 | ↓Expression | GSTA1 is involved in glutathione conjugation of the active metabolites of cyclophosphamide (CPA). GSTA1∗B allele is associated with higher survival rate of breast cancer patients treated with CPA-containing chemotherapy. | |
| GSTM1∗0 | Gene deletion | 42-58 | 27-41 | Abolished activity | The GSTM null genotype is associated with an increased risk for lung, colon, and bladder cancer. | |
| AML patients carrying GSTM∗0 appear to have a better response to adriamycin and cyclophosphamide treatment. | ||||||
| GSTP1∗B | I105V | 6-40 | 54 | ↓Activity | The GSTP1∗B allele is associated with lower clearance of etoposide and reduced risk for relapse in childhood ALL patients. | |
| The GSTP1∗B allele is associated with increased survival rate in patients with advanced colorectal cancer or breast cancer. | ||||||
| GSTT1∗0 | Gene deletion | 2-42 | Abolished activity | The GSTT1 deletion is associated with reduced risk for relapse in childhood ALL patients. | ||
| The GSTT1 deletion is a poor prognostic factor for survival in adult ALL. | ||||||
| NAT d | NAT1∗14 and ∗17 are associated with slow acetylator phenotype. | |||||
| NAT1∗4 | Wild-type | Normal | NAT2∗5, ∗6, ∗7, ∗10, ∗14, and ∗19 lead to slow acetylator phenotype. | |||
| NAT1∗14 | R187Q | 1.3-3.7 | ↓Activity | NAT2 slow acetylator phenotype is associated with increased susceptibility to hydralazine- and isoniazid-induced toxicity. | ||
| NAT1∗14 | R187Stop | ↓Activity | NAT2 slow acetylator phenotype is associated with increased risk for bladder cancer. | |||
| NAT1∗17 | R64W | ↓Activity | ||||
| NAT1∗19 | R33Stop | ↓Activity | ||||
| NAT1∗22 | D251V | ↓Activity | ||||
| NAT2∗4 | Wild-type | Normal | ||||
| NAT2∗5 | I114T | ↓Activity | ||||
| NAT2∗6 | R197Q | ↓Activity | ||||
| NAT2∗7 | G286E | ↓Activity | ||||
| NAT2∗10 | E167K | ↓Activity | ||||
| NAT2∗14 | R64Q | ↓Activity | ||||
| NAT2∗17 | Q145P | ↓Activity | ||||
| NAT2∗19 | R64W | ↓Activity | ||||
| TPMT e | TPMT is involved in the methylation reaction of mercaptopurine, an anticancer drug used in the treatment of childhood ALL. | |||||
| TPMT∗2 | A80P | 0-0.5 | 0 | 0-0.4 | ↓Activity | The TPMT genotype correlated well with in vivo enzyme activity and is clearly associated with a risk for mercaptopurine-induced toxicity. Patients with poor or intermediate TPMT activity may tolerate only one-tenth to half of the average mercaptopurine dose. |
| TPMT∗3A | A154Y, Y240C | 0-0.6 | 0-1 | 0-0.8 | Abolished activity | |
| TPMT∗3B | Y240C | – | 0 | – | 9-fold ↓Activity | |
| TPMT∗3C | A154Y | 0.2-3.3 | 0-0.2 | 2.4-7.6 | 1.4-fold ↓Activity | |
a Data on UGT SNP allele frequencies, function effect, and clinical relevance are summarized from , .
b Data on SULT1A1 SNP allele frequencies, function effect, and clinical relevance are summarized from , .
c Data on GST SNP allele frequencies, function effect, and clinical relevance are summarized from , .
d Data on NAT SNP allele frequencies, function effect, and clinical relevance are summarized from , , .
e Data on TPMT SNP allele frequencies, function effects, and clinical relevance are summarized from , .
The human UGT superfamily is a group of conjugating enzymes that catalyze the transfer of the glucuronic acid group of uridine diphosphoglucuronic acid to the functional group (e.g., hydroxyl, carboxyl, amino, sulfur) of a specific substrate ( ). UGTs are membrane-bound enzymes localized in the endoplasmic reticulum of liver and many other extrahepatic tissues. Seventeen human UGT genes have been identified and are classified into two subfamilies ( UGT1 and UGT2 ). Glucuronidation increases the polarity of the substrates and facilitates their excretion in bile or urine. Genetic polymorphisms have been identified for almost all the UGT family members. These genetic variations may alter the function or expression of the enzyme, and consequently modify the glucuronidation capacity of the enzyme toward the substrates. There is evidence that genetic variations in the UGT genes contribute to differential susceptibility to diseases (e.g., cancer) and influence the pharmacokinetics and clinical outcome of substrate drugs (for reviews, see and ). A representative example is that the UGT1A1 low promoter activity alleles (i.e., UGT1A1∗28 ) are associated with decreased glucuronidation of SN-38 (an active metabolite of irinotecan), thereby leading to increased risk for irinotecan-induced toxicity. The pharmacogenetics of irinotecan is discussed in detail later. The most common functional polymorphisms in the major UGT enzymes and their clinical relevance are summarized in Table 75.2 (for comprehensive reviews, see and ].
Cytosolic SULTs are phase II enzymes that catalyze the transfer of the sulfonyl group from the cofactor 3′-phosphoadenosine 5′-phosphosulfate to the nucleophilic sites of a variety of substrates, including hormones and xenobiotics. Sulfo conjugation of xenobiotics leads to formation of polar, excretable products as well as reactive, potentially mutagenic and carcinogenic metabolites ( ). Eleven SULT proteins encoded by 10 genes have been identified in humans. They differ in substrate specificity and tissue distribution. Single nucleotide polymorphisms (SNPs) have been identified in most of the human SULT genes. Functional SNPs in SULTs, which result in altered enzymatic activity, have the potential to influence therapeutic response and modify cancer susceptibility (see Table 75.2 ) (see reviews in and ). A most commonly occurring SNP, SULT1A1∗2 (Arg213His), exhibits reduced enzymatic activity and thermal stability.
The superfamily of human GST catalyzes the conjugation of glutathione (GSH) to a large variety of endogenous metabolites and xenobiotics, including alkylating and free radical–generating anticancer drugs such as cyclophosphamide, anthracyclines, and topoisomerase II inhibitors ( ). Human GSTs are categorized into three main families: cytosolic/nuclear, mitochondrial, and microsomal GSTs. The cytosolic GSTs are further divided into seven classes: alpha, mu, omega, pi, sigma, theta, and zeta. Their typical function is to detoxify reactive metabolites, while they may also play a role in the formation of cytotoxic metabolites ( ). Besides enzymatic function, GSTs also possess nonenzymatic functions, in which they act as regulators of cell signaling ( ; ) and posttranslational modification pathway in response to stress, growth factors, and deoxyribonucleic acid (DNA) damage, and in cell proliferation, cell death, and other processes that ultimately lead to tumor growth and drug resistance. These multiple functionalities establish the importance of GSTs as determinants of cancer susceptibility and prognosis, as well as therapeutic response (see reviews in and ). The human genes encoding GSTs are highly polymorphic. A homozygous deletion (GST-null genotype) of GSTM1 or GSTT1 occurs in about 50% or 25%, respectively, of most populations ( ). GST polymorphisms have been linked to cancer incidence, treatment outcome, and prognosis ( Table 75.3 ) (for comprehensive reviews, see and ).
| Gene | Protein | Tissue Distribution | Polarity | Representative Drug Substrates |
|---|---|---|---|---|
| ABC Transporters | ||||
| ABCB1 | MDR1 (P-gp) | Liver, intestine, kidney, blood-brain barrier, lymphocytes, placenta | AP | Anthracyclines, taxanes, vinca alkaloids, imatinib, etoposide, levofloxacin, erythromycin, cyclosporine, tacrolimus, digoxin, quinidine, verapamil, diltiazem, ritonavir, saquinavir, talinolol, phenytoin, cimetidine, simvastatin, morphine, hydrocortisone |
| ABCC1 | MRP1 (GS-X) | Ubiquitous | BL | Anthracyclines, vinca alkaloids, irinotecan, SN-38, methotrexate, camptothecins, saquinavir, ritonavir, difloxacin, drug-glucuronate/-glutathione/-sulfate conjugates |
| ABCC2 | MRP2 (cMOAT) | Liver, kidney, intestine | AP | Anthracyclines, vinca alkaloids, methotrexate, camptothecins, rifampin, pravastatin, and drug-glucuronate/-glutathione/-sulfate conjugates |
| ABCG2 | BCRP | Liver, intestine, placenta, breast | AP | Anthracyclines, irinotecan, SN38, SN38G, imatinib, tamoxifen |
| SLC Transporters | ||||
| OATP Family | ||||
| SLC21A3 | OATP1A2 (OATP-A) | Ubiquitous, with highest expression in brain and testis | BL | Rosuvastatin, methotrexate, ouabain, d -penicillamine |
| SLC21A6 | OATP1B1 (OATP-C) | Liver | BL | Statins, pravastatin, fexofenadine, repaglinide, rosuvastatin, ouabain, d -penicillamine, rifampin |
| SLC21A8 | OATP1B3 (OATP8) | Liver | BL | Digoxin, rifampin, ouabain, methotrexate, d -penicillamine, rosuvastatin, cyclosporine |
| SLC21A9 | OATP2B1 (OATP-B) | Ubiquitous | BL | Benzylpenicillin, rosuvastatin |
| OCT Family | ||||
| SLC22A1 | OCT1 | Liver | BL | Metformin, cisplatin, oxaliplatin, imatinib, procainamide, citalopram, cimetidine, quinidine, verapamil, acyclovir |
| SLC22A2 | OCT2 | Kidney | BL | Metformin, cisplatin, oxaliplatin, imatinib, procainamide, citalopram, cimetidine, quinidine, amantadine |
| SLC22A3 | OCT3 | Brain, liver, kidney, heart, muscle, placenta, and blood vessels | BL | Cimetidine, agmatine, adefovir, catecholamines |
| OAT Family | ||||
| SLC22A6 | OAT1 | Kidney, brain | BL | Methotrexate, salicylate, antiviral agents (e.g., acyclovir) |
| SLC22A7 | OAT2 | Liver, kidney, | BL | Methotrexate, salicylate, tetracyclines |
| SLC22A8 | OAT3 | Kidney, brain, muscle | BL | Methotrexate, antiviral agents (e.g., acyclovir), cimetidine, pravastatin, salicylate |
| SLC22A11 | OAT4 | Kidney, placenta | AP | Methotrexate, cimetidine, salicylate, tetracyclines |
The human NATs catalyze the transfer of an acetyl group from acetylcoenzyme A to arylamines, arylhydroxylamines, and arylhydrazines ( ). The two human NAT genes, NAT1 and NAT2, carry functional polymorphisms that influence the enzyme activity. Based on the level of NAT activity, patients can be classified into two phenotypes: fast acetylator (wild-type NAT acetylation activity) and slow acetylator (reduced NAT enzyme activity). For example, polymorphisms or haplotypes in NAT1 (e.g., NAT1∗14, ∗15, ∗17, ∗19, and ∗22 ) and NAT2 (e.g., NAT2∗5, ∗6, ∗7, ∗10, ∗14, and ∗17 ) lead to slow acetylation phenotype (see Table 75.3 ) (for a review, see ). NAT2 plays an important role in the activation and/or deactivation of a large and diverse number of aromatic amine and hydrazine drugs used in clinic, and therefore the NAT2 genotype is particularly relevant to the response to these drugs. One representative example is the association of the NAT2 slow-acetylator phenotype with increased risk for an antituberculosis drug (isoniazid)–induced hepatitis ( ). In addition, because NAT1 and NAT2 catalyze the bioactivation (via O -acetylation) of aromatic and heterocyclic amine carcinogens, genetic variations in NAT1/2 genes may modify the cancer risk related to exposure to these carcinogens ( ). For instance, the NAT2 slow-acetylator phenotype has been linked to a higher risk for bladder cancer, particularly in cigarette smokers ( ; ).
TPMT is best known for its key role in the metabolism of the thiopurine drugs (e.g., 6-mercaptopurine, azathiopurine, and 6-thioguanine) by catalyzing the S -methylation of thiopurine drugs via S -adenosyl- l -methionine as the S -methyl donor. These drugs are clinically used to treat cancers, in particular acute lymphoblastic leukemia (ALL), or as immunosuppressants. The TPMT gene exhibits significant genetic polymorphisms across all ethnic groups studied, with 18 TMPT alleles identified to date. The three main TPMT alleles, namely TMPT∗2 (reduced activity), ∗3A (abolished activity), and ∗3C (reduced activity), account for 80% to 95% of the intermediate and poor metabolizers (see Table 75.3 ) (for a review, see ). About 90% of individuals have two wild-type TPMT alleles ( TPMT∗1 ) (resulting in high TPMT activity); about 10% of individuals have one wild-type TPMT allele and one nonfunctional variant allele (resulting in intermediate activity); and rare (1 in 300) individuals inherit two nonfunctional variant alleles (resulting in complete TMPT deficiency) ( ). Individuals who inherit defective TPMT alleles or TPMT deficiency (i.e., two nonfunctional alleles) are at significantly increased risk for thiopurine-induced hematopoietic toxicity. Indeed, patients with absent TPMT activity (∼0.3% prevalence) or low activity (∼10% prevalence) may tolerate only 5% to 50% of the average mercaptopurine dose. Clinical diagnostic tests are now available for the detection of the SNPs in the human TPMT gene that lead to decreased or abolished enzyme activity. TPMT variant pharmacogenetic testing is recommended before treating patients with azathiopurine, mercaptopurine, and thioguanine.
In addition to drug-metabolizing enzymes, uptake and efflux transporters that facilitate the movement of drugs into and out of cells are important determinants of drug disposition and response. Broadly, drug transporters are classified into two families: efflux transporters of the adenosine-5′-triphosphate (ATP)–binding cassette (ABC) family and uptake transporters of the solute carrier (SLC) family. In the ABC transporter family, 49 genes have been identified and classified into seven subfamilies from ABCA through ABCG based on the sequence homology. The ABC transporters are responsible for transport of diverse substrates out of cells using ATP as an energy source. Among these, ABCB1, ABCC1/2, and ABCG2 have been well characterized for their roles in drug disposition and response. In the SLC family, 360 genes have been identified and classified into 46 subfamilies. Of particular relevance to drug disposition are organic anion–transporting polypeptides (OATPs), organic cation transporters (OCTs), and organic anion transporters (OATs).
The pharmacologically most important ABC transporters (including ABCB1, ABCC1/2, and ABCG2) and SLC transporters (including OATP, OCT, and OAT families), their tissue distributions, and representative drug substrates are summarized in Table 75.3 . These transporters play a crucial role in the intestinal absorption, biliary excretion, renal excretion, and tissue/cellular penetration of a wide variety of therapeutic drugs, and therefore they are important determinants of drug exposure in the system and at the site of action ( Fig. 75.3 ). Genetic polymorphisms in transporter genes may influence the expression, subcellular localization, substrate specificity, and intrinsic transport activity of transporter proteins, and consequently may influence the disposition and response of substrate drugs. The sections that follow highlight the functional and clinical significance of the most commonly naturally occurring genetic polymorphisms within the pharmacologically most important ABC and SLC transporters. A comprehensive list of genetic variants in the ABC and SLC transporters and related information are available in Pharmacogenetics Research network databases at http://www.pharmGKB.org .
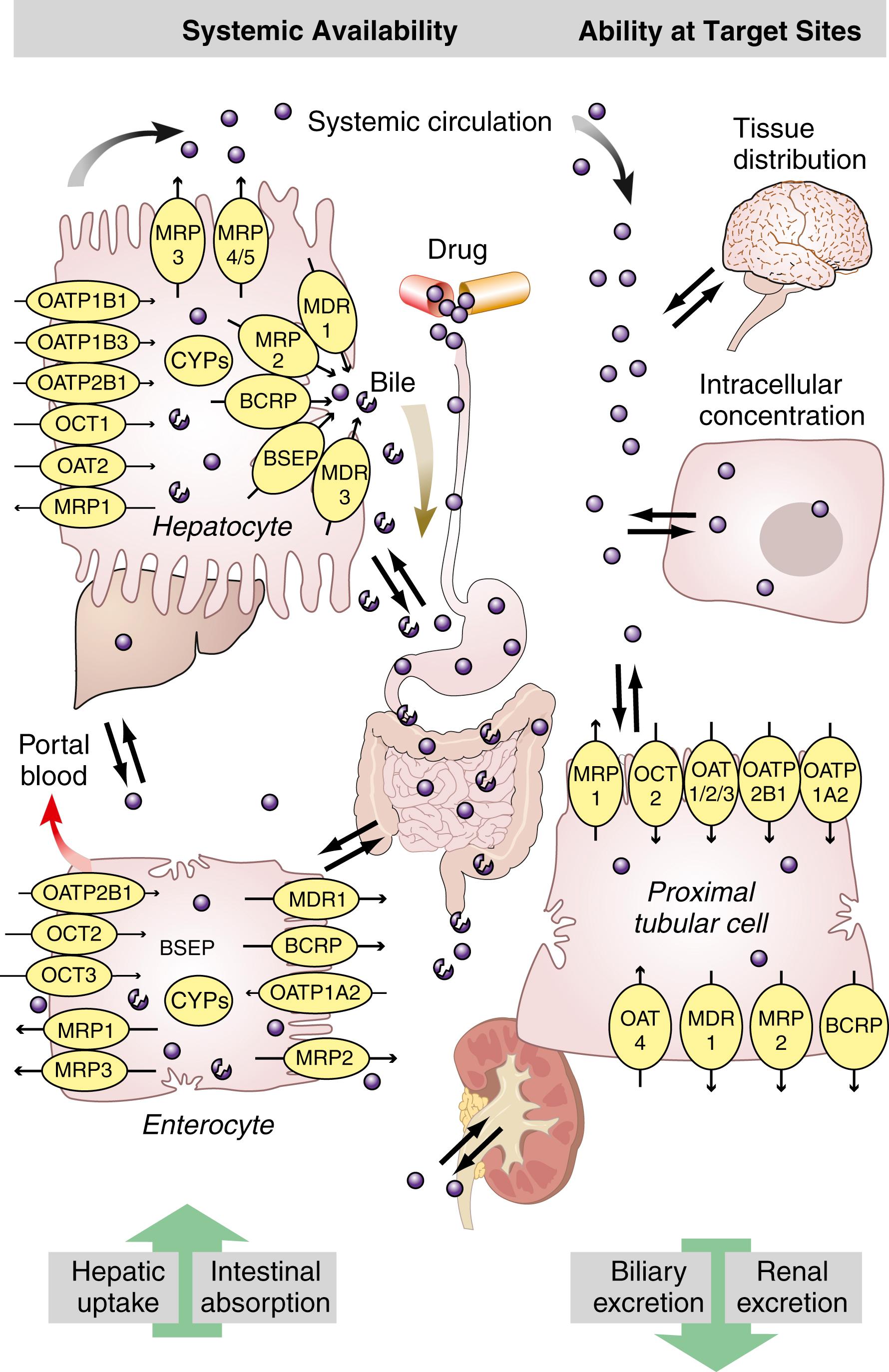
The ABCB1 gene, also named as the multidrug resistance 1 (MDR1) gene, encodes a polypeptide (P-glycoprotein) that has two halves, each containing six hydrophobic transmembrane domains and an ATP-binding domain. ABCB1 is expressed in the intestinal epithelium, canalicular membrane of hepatocyte, brush border of the renal tubule, pancreatic ductile cell, trophoblast of placenta, capillary endothelial cells of brain and testes, and peripheral blood lymphocytes (see Table 75.3 ). ABCB1, located on the apical or luminal surface of the epithelial cells, functions as an efflux transporter in restricting intestinal absorption, facilitating hepatobiliary excretion and renal excretion, and protecting the brain and fetus from xenobiotics (see Fig. 75.3 ). In addition, ABCB1 overexpression in cancer cells is implicated in multidrug resistance to chemotherapeutic agents ( ). ABCB1 transports a broad spectrum of structurally and functionally diverse drugs, including anticancer agents, antibiotics, immunosuppressants, cardiac drugs, calcium channel antagonists, and human immunodeficiency virus (HIV) protease inhibitors (see Table 75.3 ). Notably, there is a large overlap in substrate specificity and tissue distribution for ABCB1 and CYP3A4/5 ( ).
More than 50 SNPs have been identified in the human ABCB1 coding region. The most common SNPs are the synonymous 1236C>T and 3435C>T and the nonsynonymous 2677G>T (Ala899Ser). The allele frequencies of these three SNPs vary in different ethnic populations ( Table 75.4 ). The 3435C>T SNP has strong linkage disequilibrium with other SNPs in the ABCB1 gene, creating common haplotypes consisting of 3435C>T combined with 2677G>T and/or 1236C>T .
| Allele Variants | Polymorphism/Substitution | ALLELE FREQUENCY (%) ∗ | Functional Effects | ||
|---|---|---|---|---|---|
| Ca | As | Af | |||
| ABCB1 | |||||
| 1236C>T | Silent | 34-42 | 60-72 | 15-21 | Affects cotranslational folding in nearby amino acids that are essential for ATP-binding and ATP hydrolysis ( ) |
| 2677G>T/A | A893S/T | 38-47/1-10 | 32-62/3-22 | 15/ND | Affects ABCB1 expression or function, but data are inconsistent ( ) |
| 3435C>T | Silent | 48-59 | 37-66 | 10-27 | Affects cotranslational folding in nearby amino acids, thereby altering substrate specificity ( ) |
| ABCB1∗13 | 1236C>T/2677G>T/3435C>T haplotype | 23-42 | 28-56 | 4.5-8.7 | Affects the inhibition of ABCB1 by a small subset of modulators ( ) |
| ABCC1 | |||||
| 128G>C | C43S | 1 | Reduced plasma membrane localization, ↓vincristine resistance in transfected cells ( ) | ||
| 1299G>T | R433S | 1.4 | Changes in transport and resistance ( ) | ||
| 2012G>T | G671V | 2.8 | Associated with anthracycline-induced cardiotoxicity ( ) | ||
| ABCC2 | |||||
| 1271A>G | R412G | DJS; ↓ in methotrexate elimination ( ) | |||
| 1249G>A | V417I | 22-26 | 13-19 | 14 | Changes in ABCC2 expression and localization ( ; ; ) |
| 3563T>A | V1188E | 4-7 | 1 | Associated with anthracycline-induced cardiotoxicity ( ) | |
| 4544G>A | C1515Y | 4-9 | Associated with anthracycline-induced cardiotoxicity ( ) | ||
| ABCG2 | |||||
| 34G>A | V12M | 2-10 | 15-18 | 4-6 | Changes in transport and resistance ( ; ) |
| 376C>T | Q126stop | 0 | 0.9-1.7 | 0 | Loss of transport activity ( ) |
| 421C>A | Q141K | 9-14 | 27-35 | 1-5 | Affects the ATP-binding domain, thereby leading to reduced transport activity ( ; ) |
∗ Data of allele frequencies are obtained from Marzolini C, Paus E, Buclin T, et al.: Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance, Clin Pharmacol Ther 75:13–33, 2004; Gradhand U, Kim RB: Pharmacogenomics of MRP transporters (ABCC1-5) and BCRP (ABCG2), Drug Metab Rev 40:317–354, 2008.
Given the important role of ABCB1 in drug absorption and disposition, genetic polymorphisms in the ABCB1 gene may influence the outcome of pharmacotherapy. The first investigation of the functional and clinical effect of ABCB1 polymorphism was reported in 2000 for a silent SNP 3435C>T , which was associated with decreased duodenal expression of ABCB1 and increased digoxin plasma concentration after oral administration in humans ( ). In the past decade, a number of preclinical and clinical studies have been conducted investigating association of the ABCB1 genotype with its tissue expression and function, as well as with the pharmacokinetics and pharmacodynamics of a wide variety of substrate drugs (see Table 75.4 ) (for a review, see ). However, data reported on the functional and clinical impact of ABCB1 polymorphisms are often inconsistent (for reviews, see , , , , and ). The discrepancy could be due to the lack of standardized methodology and assays among different studies. In addition, a particular SNP may often result in a very subtle functional outcome. It has been shown that the 3435C>T SNP affects the timing of cotranslational folding and insertion of ABCB1 into the membrane, thereby altering substrate specificity ( ). Interestingly, the 1236C>T-2677G>T-3435C>T haplotype did not result in a change in substrate transport per se but influenced the inhibition of transport by a small subset of modulators ( ). Furthermore, conflicting results could be attributable to confounding factors from other transporters or metabolizing enzymes given the multiple disposition pathway of the substrate drug. For example, the commonly used in vivo ABCB1 probe drugs (e.g., digoxin, fexofenadine, and talinolol) are the dual substrates for both ABCB1 and OATP transporters; cyclosporine is not only transported by ABCB1 but also metabolized by CYP3A4. Thus the impact of ABCB1 polymorphisms on the pharmacokinetics of these substrate drugs could be obscured by the activity of OATP or CYP3A4. Hence, a systemic analysis of polymorphisms in multiple genes known or suspected to contribute to drug disposition and response will be essential to better understanding of the genetic impact on pharmacotherapy. In addition, the ABCB1 has multiple polymorphisms, some of which are in linkage disequilibrium, and therefore a haplotype approach would allow a more accurate prediction of clinical phenotypes.
ABCC1/2, also called multidrug resistance-related proteins, play an essential role in transport and excretion of organic anions, including physiologic metabolites, carcinogens, and drugs. They also contribute to multidrug resistance to chemotherapeutic agents ( ). ABCC1 and ABCC2 have overlapping substrate specificities, typically glutathione, glucuronate, and sulfate conjugated and unconjugated drugs, including many anticancer agents (e.g., vincristine and doxorubicin), HIV protease inhibitors (e.g., ritonavir and saquinavir), and antibiotics (e.g., difloxacin and grepafloxacin) (see Table 75.3 ). Both ABCC1 and ABCC2 require cotransport of reduced glutathione to transport some of their substrates ( ). ABCC1 is located in basolateral membranes of polarized cells, whereas ABCC2 is located in the apical domain. While ABCC1 is ubiquitously expressed, ABCC2 is mainly expressed in the liver, renal proximal tubules, intestine, and brain (see Table 75.3 ).
The human ABCC1 gene appears to be a conserved gene because many of the naturally occurring genetic variants in ABCC1 are relatively rare. Of the identified SNPs in the noncoding and coding region of ABCC1, 16 are known to result in amino acid changes, and some of them exhibit functional effects on either expression or function of the protein (see Table 75.4 ) (for a review, see ). However, data regarding the impact of ABCC1 polymorphisms on in vivo physiology, clinical drug resistance, or toxicity are rather limited. Notably, one study has identified significant associations of ABCC1 2012G>T (Gly671Val) and a haplotype of ABCC2 with anthracycline-induced cardiotoxicity among patients with non-Hodgkin lymphoma treated with doxorubicin ( ).
Mutations in the ABCC2 gene have been initially identified in Dubin-Johnson syndrome, a relatively rare recessive disorder characterized by conjugated hyperbilirubinemia resulting from loss of expression and function of ABCC2 in the liver (see Chapter 22 ). However, the impact of this loss of hepatic ABCC2-mediated transport on the pharmacokinetics of substrate drugs in humans remains to be determined. Among more commonly occurring ABCC2 SNPs, the most widely studied is 1249G>A (Val417Ile). The effect of this SNP on ABCC2 expression is different depending on the tissue examined. For example, 1249G>A was associated with lower ABCC2 mRNA and protein levels in preterm placenta, but not in duodenum and liver ( ; ). Of note, a possible association of 1249G>A variant with tenofovir-induced renal proximal tubulopathy has been demonstrated, suggesting this SNP may influence renal excretion of some ABCC2 substrates ( ). In addition, 1249G>A has been associated with the change of ABCC2 localization in neuroepithelial tumors ( ). A number of other nonsynonymous and synonymous SNPs have been studied for their potential functional influence on ABCC2 expression and transport activity (see Table 75.4 ). ABCC2 SNPs appear to have varying effects on different organs or substrates, or between in vitro and in vivo studies (for a review, see ).
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here