Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Over the past two decades, discoveries in basic immunology and the pathogenesis of malignancies have significantly advanced our understanding of the origin of lymphoid neoplasms. These diseases have been reexamined and grouped based on recurrent chromosomal rearrangements, histologic patterns, gene expression profiles, and more recently by patterns of gene mutations, gene fusions, and epigenetic alterations. The multiple revisions to the World Health Organization’s classification schemes for lymphomas reflect our evolving understanding of groupable lymphoma subtypes. As with other cancers, lymphoma development is dependent upon acquisition of mutations, DNA copy number changes, recurrent cytogenetic rearrangements, and epigenetic dysregulation of gene expression involving oncogenic and tumor suppressor pathways. Many of these derangements occur as a result of disordered genetic recombination and somatic hypermutation (SHM) events intended to support adaptive immunity. Also, analysis of molecular features of lymphomas compared to normal lymphocyte compartments provides clues to the events driving their pathogenesis.
An evolving understanding of the steps involved in lymphoid development has provided further insights into the development of this diverse group of malignancies because most non-Hodgkin lymphomas (NHLs) reflect stages of lymphoid development. The application of gene expression profiling (GEP) and massively parallel high-throughput sequencing, as well as a better appreciation of the contribution of microRNAs (miRNAs) and epigenetic alterations to lymphoma pathogenesis, have shed new light on the mechanisms underlying lymphomagenesis. Understanding of clinical and epidemiologic features of non-Hodgkin lymphomas provides additional insight into the origin of these malignancies. In this review, we emphasize that a collaboration of genetic alterations (some of which are induced by the unique biology of the adaptive immune system) collaborated with host features including immune stimulation and lack of tumor surveillance to drive the formation of lymphomas that mirror the phenotypic characteristics of their underlying cell of origin (this can be conceptualized as in Fig. 81.1 ).
This chapter reviews the most common NHL subtypes, focusing on common B-cell lymphoma subtypes, with regard to classification using prevailing views regarding genetic or genomic classification of these disorders. Additionally, insights regarding pathogenesis, prognosis, and possible therapeutic targets gleaned from genomic approaches are discussed.
Greater than 85% of NHL cases have a B-cell phenotype. With rare exception, B-cell lymphomas represent immortalized, “frozen” stages of B-cell development and the underlying molecular features of B-cell tumors reflect biologic features found in analogous normal B-cell compartments. Understanding that regulation of B-cell compartments that mirror B-cell NHL subtypes is thus crucial to understanding the driving forces behind lymphoma development ( Fig. 81.2 ).
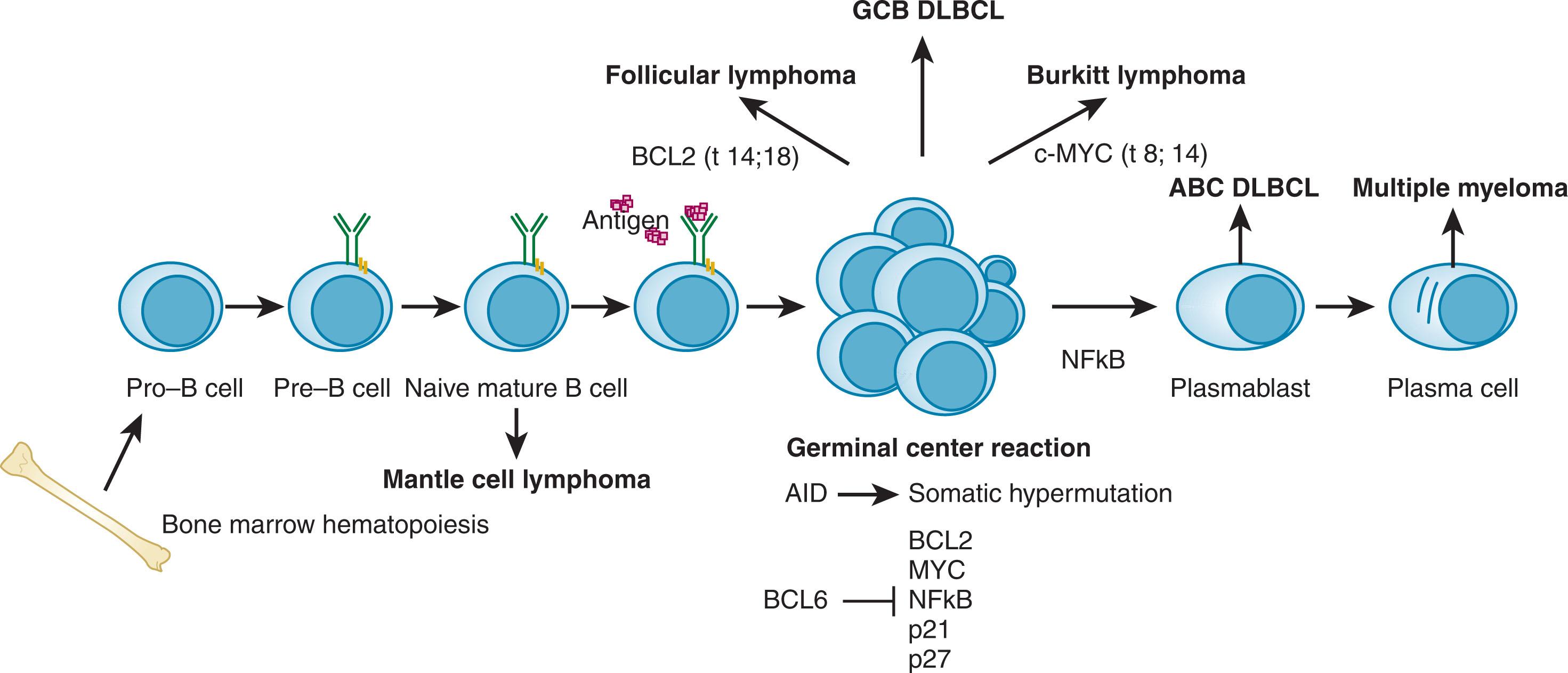
The goal of early B-cell development is to produce mature B cells expressing surface immunoglobulin M (IgM) that can efficiently recognize cognate antigen. After this is accomplished, B cells develop along pathways involving germinal center (GC), mantle, and marginal zones in lymphoid organs where they encounter antigen in association with activated antigen-presenting cells (APCs). Differentiation of mature B cells into plasma cells after SHM is the final step in B-cell development and is critical to the development of effective humoral immunity. Antibody diversity is accomplished through the process of somatic V(D)J recombination at the pro–B-cell stage via recombinase activating genes ( RAG1 and RAG2 ) followed by trafficking to GCs, where they proliferate rapidly with dividing times of 6 to 8 hours. High-affinity B-cell clones produced in part via activation-induced cytidine deaminase (AID) expression and SHM are favored because of protection from apoptosis. Through this mechanism, protective antibodies are produced at the risk of exposing antigen-stimulated B cells to genetic lesions caused by the molecular machinery utilized to create V(D)J recombination and SHM. Many NHLs exhibit translocations of driver oncogenes to highly active Ig loci, implying that recombination events intended to produce high-affinity antibodies are misdirected and instead produce immortalized B cells representing stages of B-cell development. Furthermore, mutations in oncogenes or tumor suppressors frequently occur in a pattern consistent with SHM at these loci. When considered together with the recurrent chromosomal rearrangements noted in NHL, this confirms the notion that these neoplasms stem from mature B cells that have entered (or are poised to enter) GCs.
Subsets of NHL recapitulate this pattern of normal B-cell differentiation at the histologic, molecular, and genomic levels and serve as a template for the classification of these diseases. This template can be applied to the entire spectrum of B-cell neoplasms, suggesting that NHLs form as a result of genetic alterations developed along this process. B-cell differentiation must be carefully choreographed at the molecular and genomic level because the expression of the RAG genes and AID cause DNA strand breaks and put the nascent lymphocyte at risk of oncogene overexpression and tumor suppressor deletion. Moreover, a coordinated regulation of gene expression is required to allow such genomic revision without triggering reflexive, protective apoptotic pathways and to allow appropriate B-cell differentiation. GC B cells highly express BCL6, and its functions are critically important to regulation of cell survival and differentiation. Targets of BCL6 also include cell cycle regulators (p21, p27) and TP53, which may overall work to facilitate cell cycle progression in the face of ongoing AID-mediated DNA strand breaks. BCL6 also represses PRDM1 /Blimp1 and serves to prevent plasmacytic differentiation. Plasma cell differentiation is mediated by upregulation of IRF4 and nuclear factor kappa-B (NFκB), which establish characteristic regulatory programs.
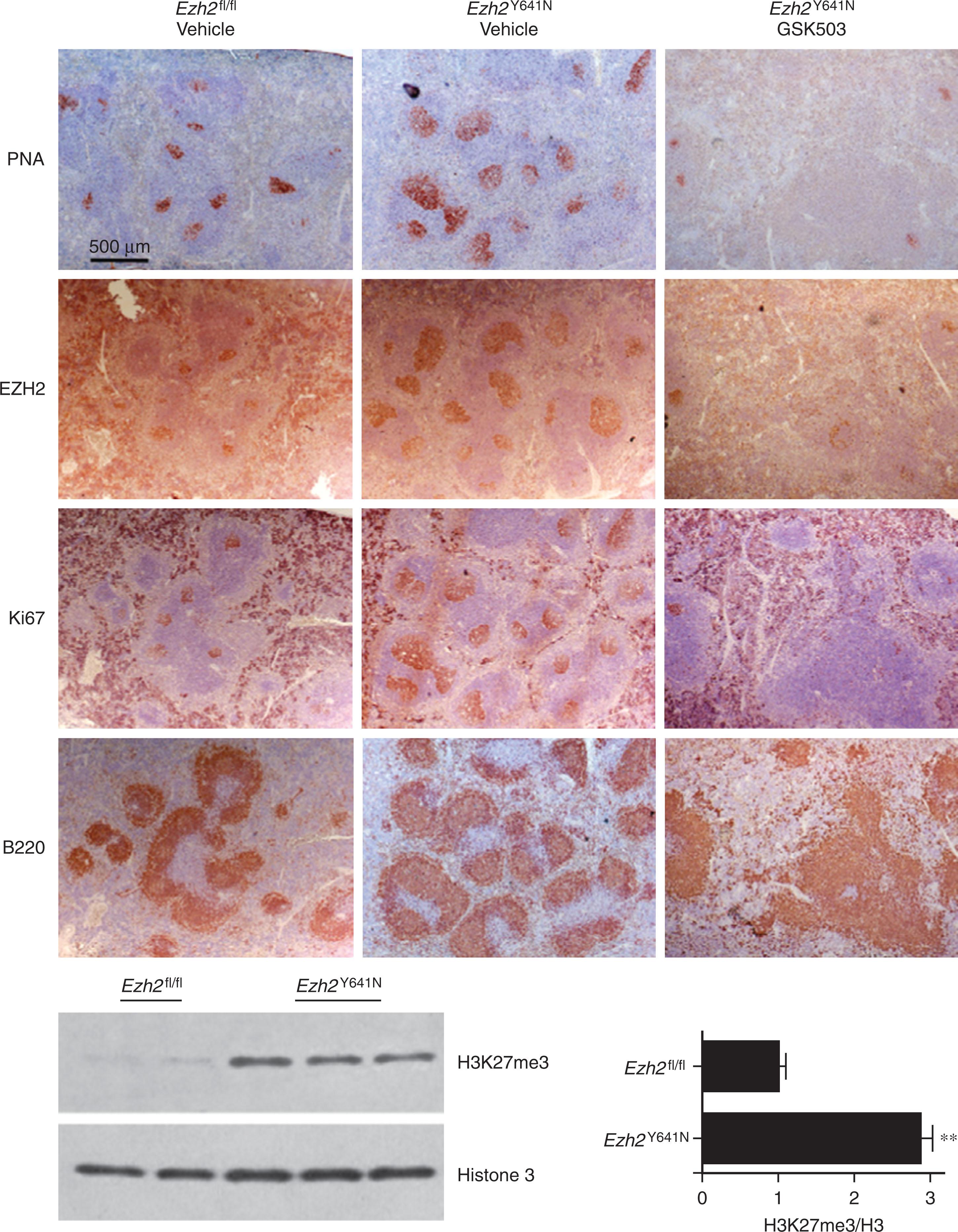
Genetic lesions altering these pathways are found recurrently in NHL. BCL6 rearrangements occur in diffuse large B-cell lymphoma (DLBCL) and other lymphomas, and constitutively active NFκB signaling is associated specifically with aggressive phenotypes of DLBCL. Translocations and or mutations of BCL2 , which is expressed at very low levels in GCs, occur in almost all follicular lymphomas (FLs) and illustrate the dangers of aberrant recombination events in the GCs because BCL2 translocation appears to be a primary event in the formation of these tumors. Recent analyses of whole genome/exome/transcriptome sequencing data have confirmed the frequent presence of these genetic lesions and have illuminated other pathways driving lymphomagenesis. In GC-derived B-cell lymphomas, mutations in EZH2 , a member of the polycomb group (PcG), occur frequently. In these tumors, EZH2 mutations result in aberrant histone methylation with resultant dysregulated control of gene expression and result in GC hyperplasia in mouse models. This process appears to enhance lymphomagenesis due to an acquired inability of GC B cells to differentiate and appropriately complete maturation and thus become transformed in a state of reflecting GC B cells ( Fig. 81.3 ). A similar role for GC regulation has been defined for genes in the sphingosine-1-phosphate receptor2 (S1PR-2) signaling pathway. Genes encoding mediators of this pathway such as S1PR2 , P2RY8 , and GNA13 are frequently mutated in GC B-cell–derived lymphomas and result in dissemination of GC cells within lymph nodes. Thus, derangements in several different genes can result in abnormal GC homeostasis and drive lymphoma formation; this is a novel paradigm whereby dysregulation of B-cell differentiation leads to transformation and lymphoma.
Regulation of antigen stimulation of the B-cell receptor (BCR) is also important in both B-cell maturation and the development of NHL. The BCR consists of a multimeric signaling complex, including CD79A and CD79B, which acts through the immunoreceptor tyrosine activation motif on CD79B and leads to a cascade of molecular events involving SYK and BTK signaling. The end result of this cascade is an increase in cellular proliferation through activation of NFκB target genes and other cellular machinery. Almost all NHLs express surface Ig, suggesting that functional BCR signaling is important at some point in the process of lymphoma formation. Analysis of Ig SHM patterns in lymphoma samples further suggests that antigen-induced selective pressure is important in the pathogenesis of NHL. The antigen(s) involved in this process are unknown, although recent reports provide evidence that BCRs on lymphoma cells may recognize other malignant cells (stimulation in an autologous fashion) or antigens within the tumor microenvironment such as host stromal tissue or chronic infection with microbes.
Epidemiologic studies have bolstered the argument that chronic immune activation and inflammation are connected with NHL development; these data also suggest that immune surveillance also plays a role in the origin of NHL. DLBCL and Burkitt lymphoma (BL) occur more frequently in immunocompromised hosts and are often Epstein-Barr virus (EBV) positive. HIV/AIDS is a significant risk factor for post-GC neoplasms, and this risk is attenuated with the use of highly active antiretroviral therapy. For example, the risk of central nervous system lymphomas is increased more than 1000-fold in patients with AIDS. NHL also occurs more frequently in acquired or inherited immunodeficiencies such as common variable immunodeficiency and severe combined immunodeficiency and with solid organ and hemopoietic transplantation. Other clinical situations in which chronic infection or inflammation occurs have also been linked to NHL. Gastric MALT (Helicobacter pylori) , hepatosplenic T-cell lymphoma (associated with infliximab use), and thyroid NHL (Hashimoto thyroiditis) are specific instances in which the development of lymphoid malignancy can be traced to chronic inflammation, either of microbial or autoimmune etiology. Finally, a critical association with EBV has been noted for posttransplant lymphoproliferative disorder, a disease that occurs in the setting of organ transplantation and immunosuppression targeting cellular immunity.
Beyond these factors, the contributions of other modifiable risk factors and heredity appear to play only a small role in NHL development. Several environmental factors (pesticides, Agent Orange, radiation exposure) have been implicated in NHL, but the associations between these agents and lymphomagenesis are difficult to prove and the effect size of these factors appears to be relatively small. Pesticide exposure does appear to have a dose-effect relationship with regard to NHL pathogenesis and has been linked to recurrent t(14;18) cytogenetic rearrangements that are noted in DLBCL and FL, but the percentage of cases linked to pesticide exposure in these entities is very low. Likewise, several genetic linkage studies have found associations between several genes and the development of various lymphoma subtypes, but the role of variations in these pathways in lymphomagenesis has not been validated. Thus, there does appear to be a small role of inherited or modifiable risk factors that likely conspire with other acquired genetic/molecular lesions in the origin of lymphoma.
And so, factors related to abnormal immune surveillance, chronic inflammation, external exposures and host factors conspire to produce genetic/molecular lesions involved in lymphoma formation and progression. The remainder of this review will focus on current knowledge of biologic features of individual NHL subtypes.
DLBCL is the most common subtype of NHL (≈25% of all NHL) and is responsible for more patient deaths than any other form of lymphoma. This entity often presents at an advanced stage, and around 50% of all patients fail to respond long-term to standard chemotherapy programs; the vast majority of those patients will eventually succumb to their disease. Even prior to the availability of GEP technology, it was clear clinically (based on tumor morphology, immunohistochemical staining, and patient outcomes) that DLBCL is a heterogeneous disease. Advances in molecular biology and genomic technologies have clarified the basis for some of this observed heterogeneity, and it has become clear that DLBCL comprises a diverse group of lymphomas that have distinct cellular origins, recurrent gene mutations, and chromosomal rearrangements that dictate their behavior. It is also clear that the stromal environment appears to play a key role in the development and progression of DLBCL, as with other cancers.
( Table 81.1 )
| Genetic Feature | ABC DLBCL | GCB DLBCL | PMBL | Notes |
|---|---|---|---|---|
| BCL2 translocation t(14;18) | 0% | 40%–50% | 20% | Alternatively translocated to light chain loci 5%–10% of time |
| BCL2 amplification | 34% | 10% | 16% | |
| BCL6 translocation t(3;V)(q27;V) | 25% | 10% | 20% | Variable translocation partners (14q32, 2p11, 22q11, 4p11, 6p21, 11q23) |
| 3q amplification | 25% | 0% | <5% | |
| 9q24 amplification | 5% | 0% | 45% | |
| PRDM1/PRDM1 deletion/mutation | 25% | 0% | Unknown | |
| REL amplification | 0 | 15% | 25% | |
| Recurrent mutation | CARD11 , MYD88 , CD79B | EZH2 , GNA13 , BCL2 | PTPN1 , SOCS1 , STAT6 | |
| MYC rearrangement t(8;14)(q24;q32) | 5%–10% of all DLBCLs | 25%–30% occur with t(14;18)(q32;q21) |
Histologic analysis and more recently high-throughput genomics assays such as GEP and massively parallel high-throughput sequencing for alterations (including somatic mutations, copy number changes, and gene fusions) have identified broad subtypes of DLBCL with molecular features that mimic their putative cell or origin; these studies have also shed light on the tremendous heterogeneity and complexity of the myriad features that drive lymphomagenesis.
GEP has identified GCB and activated B-cell–like (ABC) DLBCL subsets that express gene signatures that mimic GCs and activated B cells ( Fig. 81.4 ). Roughly 30% of DLBCLs cannot be classified using this system and have been deemed unclassified DLBCL. There are thousands of genes that differ between these subtypes at the GEP level, which is a magnitude similar to that seen between acute myeloid and lymphoblastic leukemias, which have profoundly different mechanisms of origin and natural histories.
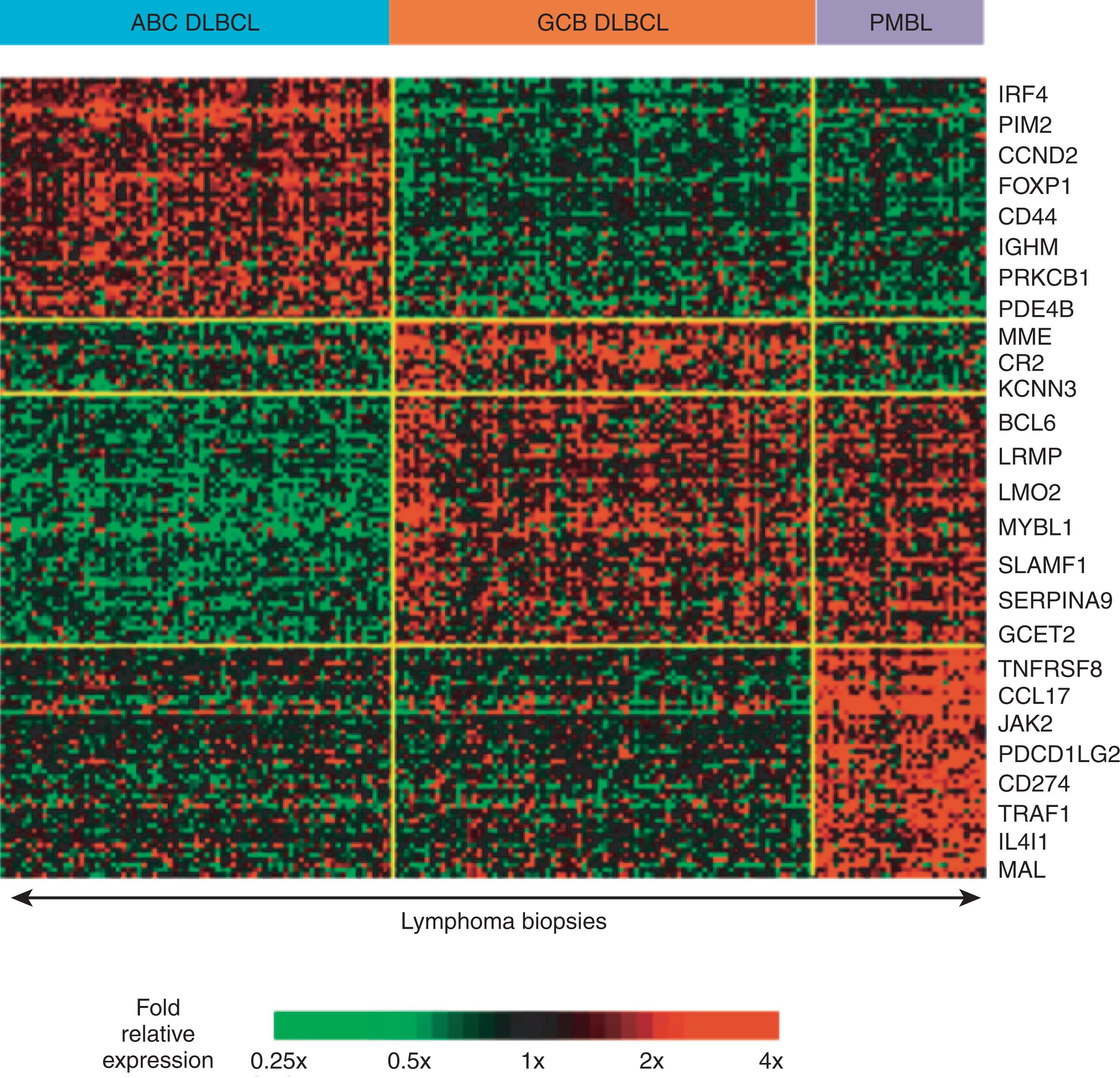
GCB DLBCLs express a characteristic spectrum of genes and have ongoing AID-mediated SHM as well as a histologic phenotype consistent with GC origin. BCL6 and LMO2 are both important to the transcriptional program of GCs and are commensurately overexpressed in GCB DLBCLs. Conversely, PRDM1 is downregulated by BCL6, which results in decreased PRDM1 activity in GCs, and GCB DLBCLs display a similar pattern. Histologically, GCB DLBCLs are characterized by CD10 expression and lack of IRF4 expression. Evaluation of Ig loci within GCB DLBCLs has also revealed evidence of ongoing SHM, which is further evidence that GCB DLBCLs reflect the biology of normal GC B cells and that dysregulation of pathways involved in GC formation might lead to this subset of DLBCL.
Alternatively, the ABC subtype of DLBCL expresses a post-GC phenotype but appears trapped just before differentiation into plasma cells. ABC DLBCLs are characterized by downregulation of many GC genes, including BCL6 and LMO2 , and by expression of genes associated with activated B cells and plasma cells. One of the hallmarks of the ABC DLBCL phenotype is upregulation of NFκB. NFκB is a transcription factor that targets a number of gene programs governing cell proliferation, immortality, and angiogenesis within NHLs. The effectors of NFκB signaling exist in inactive form in the cytosol and require signaling either through MAPK/ERK (mitogen-activated protein kinase/extracellular signal-regulated kinase) signaling (the canonical NFκB pathway) or through ligation of CD40–LTbR–BAFF-R (B-cell activating factor receptor), which results in phosphorylation of the inactive complex and translocation of either the p50–RelA or p52–Rel B complexes to the nucleus and subsequent transcription of target genes. In ABC DLBCL cell lines, a signaling complex involving CARD11, BCL10, and MALT1 (the CBM complex) constitutively activates NFκB signaling and selective knock-down of any of the components of this complex using RNAi or small molecule inhibitors and is lethal to ABC cell lines. Interestingly, a few GCB cell lines also demonstrate activation of NFκB and mutations in the NFκB pathway, which might confer sensitivity to inhibition of this pathway in those tumors. The mechanisms by which DLBCLs acquire and maintain constitutive NFκB activity are still being elucidated and may provide insights into the origin and into possible treatments for this entity.
Primary mediastinal B-cell lymphoma (PMBL) represents a distinct subset of patients that have sometimes been classified as DLBCL. PMBL was originally identified based on the clinical characteristics of the disease. Patients with PMBL are generally young (third or fourth decades of life), predominantly women, and present with a large mediastinal mass; outcomes with chemoimmunotherapy for PMBL are generally superior to those with other subtypes of DLBCL. GEP studies revealed that PMBL could be clearly delineated from ABC and GCB subtypes. Indeed, by gene expression, PMBL appears similar to Hodgkin lymphoma with many shared features in both diseases, including the expression of CD30 ( Fig. 81.5 ). Both diseases also express genes associated with Janus-activated kinase (JAK)–signal transducer and activator of transcription (STAT) signaling as well as targets of the NFκB pathway.
Investigation of the molecular landscape of DLBCL has revealed a set of recurrent cytogenetic and molecular abnormalities associated with pathogenesis. Chromosomal translocations involving BCL6 , BCL2 , and MYC (v- myc myelocytomatosis viral oncogene homolog) occur frequently in DLBCL, and these translocations appear to reflect cell-of-origin phenotype. 3q27 rearrangement with Ig gene partners is the most common rearrangement seen in DLBCL and occurs in about 30% of both the ABC and GCB subtypes. This genetic rearrangement upregulates the BCL6 gene, resulting in an oncogenic phenotype. Additional mutations in the transcriptional regulatory program of BCL6 may also occur with alternative translocation. Notably, BCL6 translocations appear to not be sufficient for lymphoma formation as evidenced by mice bearing t(3;14) translocations that develop NHL at a low rate. BCL2 translocations occur in a minority of GCB DLBCLs but are not seen in the ABC phenotype; this mirrors the situation in FL, in which BCL2 overexpression results in disordered GC transit. MYC rearrangements additionally occur in DLBCL (≈5% of cases), sometimes with rearrangements of BCL2, BCL6 , or both. These tumors, sometimes termed double-hit lymphomas , display an aggressive phenotype seemingly intermediate between BL and DLBCL.
Oncogenic mutations and areas of chromosomal amplification or deletion also appear important in the pathogenesis of DLBCL. Upregulation of c-REL occurs in about 20% of GCB tumors through amplification of the 2p locus. Amplification of MYC and BCL2 may also occur through this mechanism. TP53 mutations and SHM of other loci, including PIM1, BCL6 , and MYC , also appear to contribute to lymphoma formation and progression. Mutations in CARD11 causing constitutive NFκB activation have been described in a subset of both ABC and GCB DLBCLs and appear to drive oncogenesis in these tumors. Overall, the patterns of gene mutation and recurrent cytogenetic abnormalities add further complexity to the classification of DLBCLs.
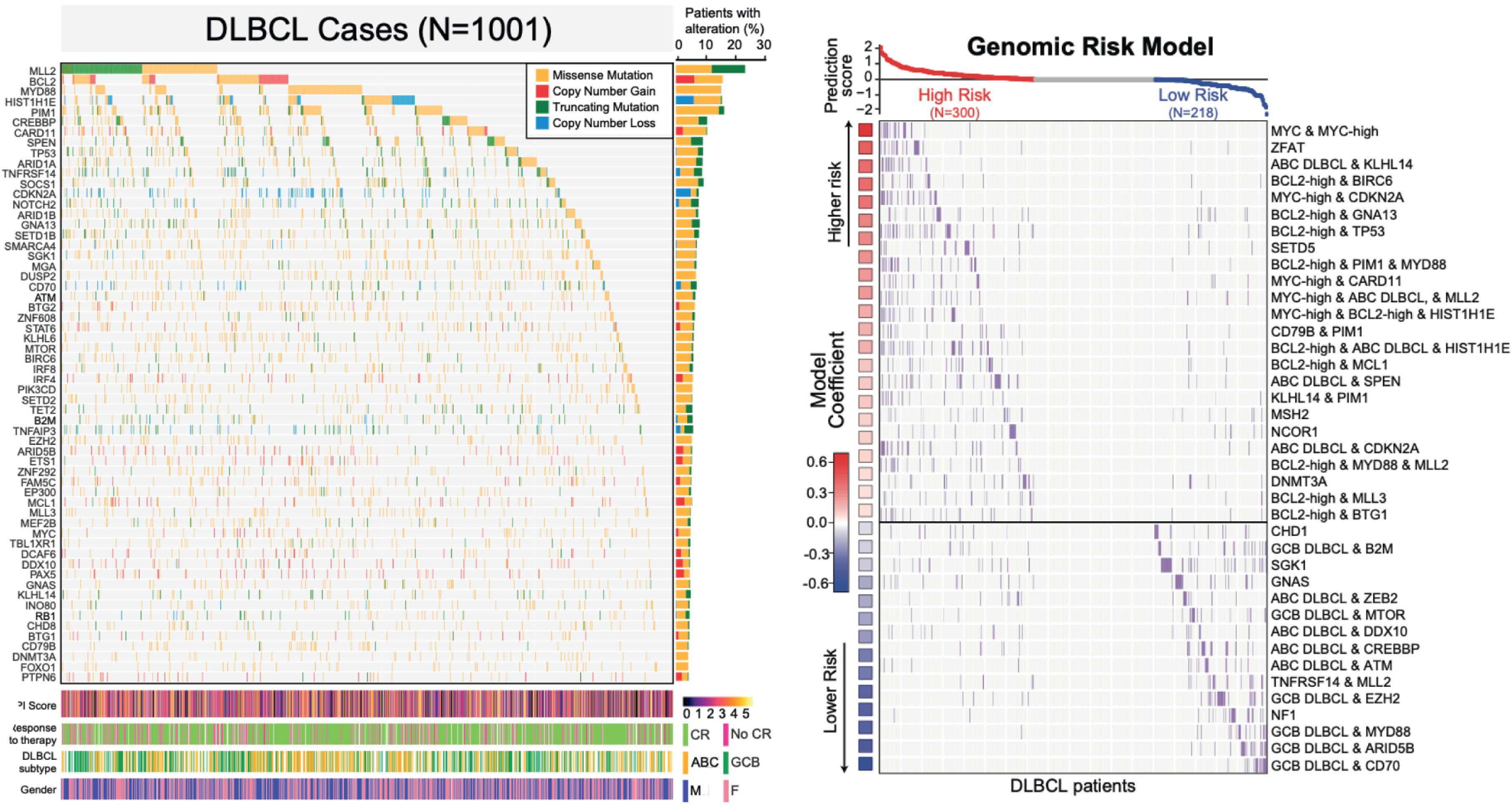
Recurrent mutations of genes involved in DLBCL pathogenesis have also been reported. Most recently, several groups have reported initial and now expanded/updated analyses of whole-genome, exome, or transcriptome data from the application of massively parallel sequencing in thousands of cases of DLBCL. DLBCL case clinical outcome data is now available in conjunction with recent data sets and allows for a refined analysis of features associated with treatment resistance and other clinical events in this entity ( Fig. 81.6 ). These studies have identified hundreds of novel mutations in DLBCL and confirm previous descriptions of survival association with cell or origin and other phenotypic features. Two of these studies show DLBCL cases can be clustered into novel subgroups based on somatic alterations including SNVs, GEP, and CNVs, although these studies have somewhat different conclusions as to molecular grouping of DLBCL cases. It may be more appropriate to consider the contribution of various mutations with pathogenesis in a combinatorial factor as there is tremendous heterogeneity in the spectrum of mutations that participate. Interestingly, some of the molecular features of lymphoma subtypes such as activation of the NFκB signaling pathway identified through GEP have been shown to correlate with specific mutations in DLBCL cancer genes. Inactivating mutations of PRDM1 and mutation in genes regulating NFκB signaling (TNFAIP3, MYD88, CARD11) have been noted in ABC DLBCL subtypes. CD79B and BCL2 have also been found to harbor mutations in DLBCL and other NHLs. A series that performed RNA sequencing on 127 NHL samples (including 83 DLBCL tumors) found 651 coding single nucleotide variants (cSNVs), many of which were not previously documented in malignancies. Truncating deletions in tumor suppression genes (TP53, TNFRSF14, CREBBP) were among the most significantly mutated genes and other heavily mutated genes not previously known to be involved in DLBCL pathogenesis (including MLL2, BTG1, EZH2 , and GNA13 ). MLL2 contained cSNVs in 26% of tumors and most often occurred as a heterozygous defect, and sometimes multiple mutations occurred in trans fashion. MLL2 functions as a H3K4-specific methyltransferase responsible for regulation of transcription of developmental genes. Ostensibly, MLL2 might serve to deregulate genes involved in differentiation or development and act in conjunction with other oncogenes to facilitate NHL. Other groups have confirmed mutations in MLL2 and other genes (CREBBP/EP300) that coordinate chromatin acetylation, leading to the hypothesis that mutations in histone-modifying genes lead to DLBCL and other NHLs. Further work is needed to dissect the mechanism(s) by which the various molecular lesions in individual tumors cause lymphoma formation and this will perhaps also improve our understanding of normal lymphocyte biology while providing new tools for lymphoma diagnosis and treatment.
The complexities of antigen presentation and recognition and regulation of B-cell proliferation that occur in GCs suggest that the microenvironment may play a role in the origin and progression of DLBCL. Tumor-infiltrating T cells have been correlated to longer survival in patients with DLBCL, and failure of immune editing is likely responsible for the increase in NHL in HIV-positive and immunosuppressed persons. GEP has been used to define stromal elements related to DLBCL progression. In one series, hierarchical clustering was used to define three groups among DLBCL samples by GEP. Clusters identifying gene signature termed oxidative phosphorylation, BCR proliferation , and host response were identified; notably, the host response group exhibited gene expression profiles associated with T and natural killer cells, dendritic cells, and macrophages. This work illustrates that patterns of stromal involvement in DLBCL could play a key role in pathogenesis. In another series involving biopsy specimens from 414 patients treated with chemotherapy and chemoimmunotherapy, two gene signatures characterizing patterns of tumor stroma best predicted survival. Improved survival was noted with a pattern that included genes associated with the extracellular matrix and histiocytic infiltration. The converse was true for gene profiles correlating to angiogenesis because these were associated with far worse overall survival.
Although it is not known what roles distinct lymph node subsets play in the pathogenesis of lymphoma, it is clear from these studies that the tumor microenvironment plays a role in the origin of NHL.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here