Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The Notch pathway is an evolutionarily conserved signaling pathway present in all metazoans that influences a wide range of developmental and physiological processes, including the maintenance of self-renewing adult cells and tissues. Since Notch is a critical regulator of proliferation and differentiation in both development and tissue homeostasis, it is not surprising that dysregulation of Notch activity or mutations within the Notch signaling pathway have been linked with inherited human disorders as well as cancer. First named after a Drosophila partial loss-of-function mutation that resulted in “irregular notches” in the wing margin, the Notch pathway has been the focus of numerous studies in worms, flies, and mammals. Canonical Notch signaling mediates direct cell-to-cell communication to establish differential cell processes in neighboring cells ( Fig. 6.1 ). Activation of the Notch pathway involves direct physical contact between cells expressing membrane-bound ligands (signal sending) and cells expressing Notch receptors (signal receiving). Ligand-binding activates sequential proteolytic processing of the Notch receptor to release the Notch receptor intracellular domain (NICD), which subsequently travels to the nucleus to activate the transcription of specific target genes. Thus, Notch signaling induces differential gene expression programs in neighboring cells. Signaling events are normally transient, with rapid degradation of NICD limiting the duration of the response. Responses are determined by the cellular context of the signaling, with NICD-targeting specific effector genes to transduce tissue-specific biological responses.
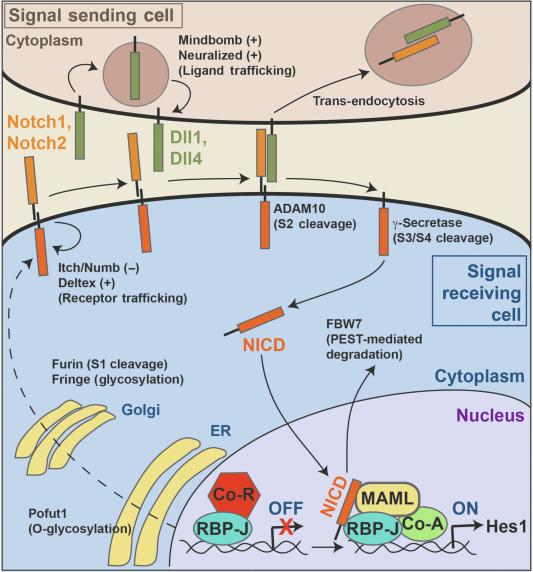
The Notch pathway therefore represents a unique mechanism for short-range cellular communication between juxtaposed cells. Developmental studies, particularly in invertebrates, have shown that this short-range signaling can function in distinct ways to regulate varied and often-divergent responses through effects on cell specification, proliferation, apoptosis, differentiation, and tissue patterning. For example, Notch signaling is involved in the process of lateral inhibition in which subtle differences in Notch signaling between two equivalent progenitor cells are transcriptionally amplified such that a bias in productive Notch signaling occurs between each cell. This unequal priming of Notch signaling leads to the establishment of each cell as either signal-sending or signal-receiving to pattern the developing tissue. Notch signaling can also occur between two distinct cell populations to establish boundary/inductive cell fate interactions associated with tissue patterning. Moreover, Notch signaling can control binary cell fate decisions between two daughter cells that are dependent on asymmetrical inheritance of Notch regulatory components. Lastly, Notch signaling has been implicated in stem cell maintenance and self-renewal through cellular interactions between stem cells and juxtaposed niche cells. In many cases of complex tissue and organ formation, these different modes of Notch signaling are used iteratively and/or in a combinatorial manner to generate complicated differentiation programs and outcomes. Importantly, Notch signaling does not act alone in these events, and invariably the Notch pathway interacts or cross-talks with other key signaling pathways (Jak/STAT, RTK, TGFβ, Wnt, BMP, Hedgehog) to establish functional and complex signaling networks required for development and tissue homeostasis. Several excellent reviews on canonical Notch signaling have been recently published. Not surprisingly, Notch signaling has been shown to play a critical role in gastrointestinal tissues. This chapter summarizes the current understanding of canonical Notch signaling mechanisms and highlights the important role that Notch plays in the intestinal epithelium to regulate stem cell self-renewal, progenitor cell proliferation, and cell fate determination.
In mammals, multiple Notch receptor and ligand family members are widely expressed during development and in adult tissues ( Table 6.1 ). The complexity of Notch receptor and ligand interactions suggests that individual ligand/receptor combinations are likely to be involved in both distinct as well as overlapping functions. Another unique feature of Notch signaling is that ligand-mediated Notch receptor proteolysis generates an active NICD fragment that does not utilize downstream secondary messengers for signal amplification. More importantly, Notch proteolysis and the initiation of Notch signaling is an irreversible process in which each activated Notch receptor can only signal once through the generation of a single NICD fragment. Thus, the availability and presentation of Notch ligands to receptors must be highly regulated, not only to control productive signaling in a spatiotemporal manner but also to prevent inappropriate signaling and the potential for oncogenic transformation. Differential expression and transcriptional regulation of Notch receptors and ligands contribute to this regulation. However, cells that express the same levels of receptors and ligands are capable of committing to different Notch-dependent cell fates, indicating that Notch activity is under additional regulation. Indeed, studies in invertebrates and mammals have highlighted the importance of posttranslational modifications and trafficking of both receptors and ligands to modulate Notch activity. The relevance of structural domains within Notch receptors and ligands in relation to key aspects of proteolysis, glycosylation, intracellular trafficking, and ubiquitin-mediated degradation that regulate Notch activity are described below ( Fig. 6.1 and Table 6.1 ).
| Component/Activity | Gene(s) a | Protein Type/Activity | Modulatory Function |
|---|---|---|---|
| Receptor | Notch1, Notch2, Notch3, and Notch4 | Type 1 transmembrane protein | |
| Ligand | Dll1, Jag1, and Jag2 | DSL and DOS domain-containing | |
| Dll3 and Dll4 | DSL only | ||
| Proteolysis | Furin-like convertase | Proprotein convertase (receptor S1 cleavage) | Heterodimer formation |
| Adam10 | Metalloproteinase (receptor S2 cleavage) | Initiates receptor signaling b | |
| TspanC8s c | Metalloproteinase trafficking/substrate presentation (receptor S2 cleavage) | Positive and negative regulators | |
| Psen(1 or 2), Ncstn, Psenen, Aph1(a, b or c) | γ-Secretase complex: 1:1:1:1 stoichiometry (receptor S3/4 cleavage) | NICD generation | |
| Adam9, Adam10, Adam12, Adam17 | Metalloproteinase (ligand) | Ecto domain shedding | |
| Glycosyltransferase | Pofut1 | O-Fucosyltransferase (receptor) | Essential for Notch activity |
| Fringe Family: Lfng, Mfng, Rfng | β1,3-GlnNAc transferase (receptor) | Optimal Notch activity/ligand specificity | |
| Poglut1 | O-Glucosyltransferase (receptor) | Essential for Notch activity Promotes extracellular cleavage |
|
| Gxylt1, Gxylt12, and Xxylt1 | O-Xylosyltransferase (receptor) | Negative regulator | |
| Eogt1 | O-GlcNAc transferase (receptor) | Impaired Notch signaling Reduced DLL1/DLL4 binding |
|
| Membrane/endosomal trafficking | Mib1 | Ring finger E3 ubiquitin ligase (ligand endocytosis) | Optimal ligand activity |
| Itch/Nedd4 family | HECT domain E3 ubiquitin ligase (receptor endocytosis) | Reduces Notch activity; lysosomal trafficking/degradation | |
| Deltex1, 2, 3, and 4 | Ring finger E3 ubiquitin ligase (receptor endocytosis) | Optimal receptor activity | |
| Numb | Receptor-binding partner (asymmetric partitioning during cell division) | Negative regulator | |
| Fbxw7 | F-Box ubiquitin ligase (NICD degradation) | Reduces Notch activity; proteosomal trafficking/degradation | |
| Crumbs | Receptor-binding partner (inhibits ligand-independent Notch endocytosis/activation) | Negative regulator | |
| Commd99 | Regulator of endosomal trafficking and Notch recycling to the cell surface | Positive regulator | |
| Ubiquitin-specific | Usp28 and Usp12 | Counteracts Fbxw7 |
a Mouse gene symbols are listed; see text for gene definitions and protein activity.
b Rate-limiting step for initiation of a Notch signaling event.
c Tspan5, 10 and 14 (positive regulators) and Tspan15 and 33 (negative regulators).
Canonical Notch signaling involves activation of Notch receptors at the cell surface by ligands of the DSL (Delta/Serrate/LAG-2) family, which includes Delta and Serrate/Jagged in Drosophila and mammals and LAG-2 in Caenorhabditis elegans. In mammals, there are four different Notch receptors (Notch1, 2, 3, 4), three Delta-like proteins (Dl1, Dll3, and Dll4), and two Jagged proteins (Jag1 and Jag2). Both Notch receptors and their ligands are type 1 transmembrane proteins. All Notch receptors consist of a multidomain protein, including an extracellular domain with 29–36 tandem epidermal growth factor (EGF)-like repeats, a heterodimer domain, a negative regulatory region (NRR) composed of three cysteine-rich LNR (Lin12, Notch repeats) repeats, a transmembrane segment, and an intracellular domain that contains the NICD signaling segment. Several of the EGF-like repeats are critical for regulating the affinity of ligand binding and signaling efficiency. For example, EGF-like repeats 11–12 are required for productive ligand signaling between neighboring cells ( trans activation), whereas EGF-like repeats 24–29, which map to the Abruptex region of Drosphila Notch, inhibit functional ligand interactions within the same cell ( cis inhibition). Recent structural analysis suggests that the alignment of Ca 2 + -binding and non-Ca 2 + -binding EGF repeats can create a hinged conformation of the Notch extracellular domain. In addition, X-ray structures of Notch1-Dll4 and Notch1-Jag1 complexes have revealed that the module at the N terminus and DSL domains of Dll4 and Jag1 interact with Notch EGF repeats 11–12 in an antiparallel fashion. It is postulated that variations to the extent of interface interactions between different Notch receptor/ligand complexes as well as ligand-binding interactions with other Notch EGF repeats may contribute to differences in ligand affinity and productive Notch activation. In the absence of ligand, the NRR domain prevents inappropriate receptor activation by masking a critical proteolytic cleavage site that only becomes accessible after ligand-induced conformational changes (see text below). The intracellular segment of the Notch receptor contains a RBP-Jκ association module (RAM) motif, ankyrin repeats, nuclear localization signals, and a proline/glutamic acid/serine/threonine-rich (PEST) motif that regulates NICD protein stability.
Classic Notch ligands (Dll1, Jag1, and Jag2) contain a DSL domain, a Delta, and OSM-11-like (DOS) domain and multiple EGF-like motifs within their extracellular domains. Additionally, both Jag1 and Jag2 contain a cysteine-rich domain located in a juxtamembrane region. Both DSL and DOS domains are involved in productive ligand binding to Notch receptors. However, similar to Notch ligands found in C. elegans , the mammalian ligands (Dll3 and Dll4) lack the DOS motif. In addition, several Notch coligands and other noncanonical Notch ligands that lack both DSL and DOS domains have been identified that act through alternative and poorly defined mechanisms and will not be discussed further.
During maturation in the secretory pathway, the Notch receptor undergoes a critical cleavage by furin-like protein convertases at site 1 (S1) to produce the extracellular N-terminal and intracellular C-terminal fragments (NTF and CTF) that form the heterodimeric receptor molecule, which is held together by noncovalent interactions ( Fig. 6.1 ). Upon ligand engagement at the cell surface, a series of receptor cleavage events in the CTF are initiated. The first proteolytic cleavage event at site 2 (S2) within the extracellular juxtamembrane region of the Notch CTF is performed by a disintegrin and metalloproteinase (Adam), which leads to removal of the Notch ectodomain and the generation of a membrane-anchored Notch CTF, termed Notch extracellular truncation (NEXT). This Adam-mediated cleavage event is the key regulatory step in the initiation of Notch signaling. Numerous studies in mice have demonstrated that Adam10 is the principal sheddase responsible for ligand-dependent Notch S2 cleavage in vivo . In addition, in vitro studies have confirmed that canonical ligand-induced proteolysis of human Notch receptors is strictly dependent upon Adam10.
The TspanC8s are members of an evolutionarily conserved tetraspanin family that interact with and promote Adam10 intracellular trafficking and enzymatic maturation. Consistent with a role of Adam10 in Notch activation, TspanC8s promote Notch activity in flies and worms. However, in mammals, different TspanC8s differentially regulate shedding of distinct Adam10 substrates, including Notch, in a manner that is likely dependent on Adam10 subcellular localization, conformation, and/or direct TspanC8 - substrate interactions.
NEXT is susceptible to subsequent sequential intramembrane proteolytic processing at sites S3 and S4 by the presenilin-dependent γ-secretase complex to release the NICD signaling domain. As part of a negative feedback regulatory mechanism, the interacting ligand (and possibly the NTF of the Notch receptor ectodomain) is trans-endocytosed into the signal-sending cell to reduce ligand availability; in addition, this may initiate other signaling events in the signal-sending cell. The γ-secretase complex is comprised of the membrane proteins presenilin (Psen), nicastrin (Ncstn), presenilin enhancer 2 homolog (Psenen), and anterior pharynx defective 1 homolog (Aph1) in a 1:1:1:1 stoichiometry. At least six possible γ-secretase complexes can be formed in rodents due to two isoforms of presenilin and three isoforms of Aph1. The biological significance of the different complex configurations is not currently known; however, biochemically, it appears that different complexes may exhibit differential processing efficiency for certain substrates and differential susceptibility to pharmacological inhibitors. In the case of ligand-dependent Notch signaling, both presenilin-1 and -2 can participate in Notch intramembrane processing leading to transcriptional activation. However, it will be important to identify the temporal and spatial expression patterns of each of the components of the γ-secretase complex in gastrointestinal tissues to determine, which complexes are formed because complex composition likely affects substrate specificity and biological function in vivo. Additionally, several membrane-anchored Notch ligands are susceptible to sequential processing by Adam and γ-secretases, but the physiological importance of these cleavage events with respect to regulation of Notch signaling remains unsolved. It is conceivable that Adam-mediated ectodomain shedding would deplete cell surface ligand levels and downregulate Notch signaling. Similarly, the production of soluble ligand could modulate and possibly interfere with productive interactions of membrane-bound ligands with cell surface Notch receptors. By contrast, intramembrane processing of Notch ligands by γ-secretases may generate intracellular fragments that have distinct signaling activities independent of Notch receptors. However, the biological significance of this alternative signaling pathway associated with Notch ligands awaits further investigation.
Following NEXT cleavage by the γ-secretase complex, the biologically active NICD is released from the membrane into the cytosol. NICD translocates to the nucleus of the Notch receptor-expressing cell and binds to the DNA-binding protein RBP-Jκ (recombination signal-binding protein for immunoglobulin kappa J region; also known as CSL), the transcriptional coactivator Mastermind (MAML) and other coactivators to initiate the transcription of target genes, including hairy and enhancer of split 1 (Hes1). In the absence of NICD, RBP-Jκ appears to occupy its DNA-binding sites some of the time and participate in a repressor complex.
The Notch signal is short lived due to rapid proteosomal degradation of NICD. The PEST motif in the C-terminus of NICD is rapidly phosphorylated by CDK8 kinase, ubiquitinated by E3 ligases such as F-box and WD-40 domain protein 7 (Fbxw7) and then sent for rapid protein degradation. Thus, study of the Notch signaling pathway is challenging because of the difficulty of detecting nuclear NICD due to its rapid degradation. As an alternative to NICD detection, assessment of a transcriptional target of Notch signaling such as Hes1 is frequently used as an indicator of Notch signaling activity; however, there are caveats to this method of detection as well because Hes1 can be regulated by other pathways, including the fibroblast growth factor pathway, GATA transcription factors, Ids, and Pax transcription factors. More recently, genetic mouse Notch reporter strains have been engineered to mark active Notch signaling at the level of sequential Notch cleavage and NICD production (e.g., NIP1-Cre) or transcriptionally using knockin CreER reporter alleles to Notch target genes, such as Hes1 and Olfm4.
Notch and DSL ligands are extensively glycosylated, with many potential sites for both N-linked and O-linked glycosylation. Studies in flies and mammals have revealed that O-linked glycosylation modifications of the Notch extracellular domain can regulate Notch activity through a variety of potential mechanisms, including modulation of ligand specificity, increasing levels of functional cell surface Notch receptors through correct folding and trafficking, and by improving ligand-mediated Notch proteolysis. The endoplasmic reticulum (ER)-localized glycosyltransferase protein O-fucosyltransferase1 (Pofut1) is required for addition of O-fucose moieties to specific Ser or Thr residues in distinct EGF-like repeat motifs of the Notch receptor. Many of the O-fucose modified EGF repeats can be further extended by the Fringe family of glycosyltransferases (discussed in more detail below). In flies and mice, analysis of loss-of-function Pofut1 mutants has revealed that this glycosylation pathway regulates Notch signaling activity. Interestingly, earlier studies in flies suggested that Pofut1 had ER chaperone activity distinct from its enzyme activity, which was required for efficient folding and transport of Notch receptors to the cell surface and optimal signaling. Similarly, in mammalian cells, overexpression of a Pofut1 mutant that lacks enzyme activity partially restored Notch signaling, suggesting that O-fucose modifications are not essential for trafficking but are needed for efficient ligand-mediated signaling. Although other unidentified ER glucosidases may contribute to Notch folding and chaperone activity, the O-fucose moieties and their subsequent modifications by Fringe glycosyltransferases are critical for Notch to signal optimally.
In mammals, three Fringe proteins, Lunatic, Manic, and Radical Fringe, are responsible for the addition of N-acetylglucosamines to O-fucosylated EGF repeats of the Notch receptor. Specifically, Fringe-mediated modifications of the highly conserved EGF-like 12 repeat alter Notch responsiveness and the binding specificity to different Notch ligands. Although Delta-like and Jagged ligands have intrinsically different Fringe-independent affinities for Notch receptors, in general, Fringe-dependent modifications enhance responsiveness of Notch receptors to Delta-like ligands over Serrate/Jagged ligands. These differences in ligand-binding affinities associated with Fringe glycosylation have been implicated in regulating the strength and location of Notch signaling involved with tissue patterning and morphogenesis in a context-dependent manner. The importance of glycosylation for the strength of Notch signaling was clearly demonstrated when a Thr-to-Ala mutation of the conserved O-fucose site within the EGF-like 12 repeat of Notch1 was introduced, and the resultant homozygous mouse mutants (Notch 12f/12f ) were viable but had reduced growth rates. Moreover, the complex expression patterns of the three mammalian Fringe proteins combined with variations and often-opposing effects on ligand responsiveness in a cell-dependent manner suggest that these glycosyltransferases may have other undefined regulatory functions.
In Drosophila , another glucosyltransferase, Rumi, has been implicated in regulating Notch signaling through the addition of O-glucose to specific serine residues of the Notch extracellular domain. These O-glycosylation modifications are not required for cell surface expression or ligand binding, but are important for correct folding and ligand (Delta)-mediated conformational changes needed for initiating Notch proteolysis and activation. A mammalian homolog of Rumi, Poglut1 can rescue Notch-phenotypes in Rumi mutant flies. Conversely, Poglut1 deletion in mice produces embryonic lethality with many Notch-like developmental defects. However, other developmental defects distinct from Notch are found in Poglut1-deficient mice suggesting that O-glycosylation is involved in regulating multiple targets critical for mammalian development. Specific xylosyltransferases, Gxylt1, Gxylt2, and Xxylt1 can further elongate O-glucose glycans through addition of xyloses. In Drosophila, xylosyl extension of O-glucose glycans on Notch inhibits Notch activation possibly by inhibiting proper Notch trafficking to the cell surface. Other O-glycosylation modifications, including O-GlcNAc glycosylation, have been detected in mammalian Notch receptors and ligands. Also in mammals, the EGF-specific O-GlcNAc transferase Eogt1 is responsible for O-GlcNAc modifications of EGF repeats. While the Drosophila homolog Eogt has been reported to have genetic interactions with Notch, the recent analysis of Eogt1-deficient mice revealed that Notch receptors lacking only O-GlcNAc glycans have reduced ligand-induced Notch signaling due to decreased affinity for Dll1 and Dll4 but not Jag1. Further studies are clearly needed to better understand which EGF repeats on Notch receptors are modified by glycosylation and how these posttranslational modifications alter Notch signaling in vivo .
Notch ligands generate productive Notch signaling through binding cell surface Notch receptors on adjacent cells ( Fig. 6.1 ). However, Notch ligand activity is critically dependent on mono-ubiquitination and endocytic trafficking, suggesting that ubiquitinated/endocytosed Notch ligands undergo posttranslational modifications upon trafficking in endocytic vesicles in order to gain biological activity. In Drosophila , two structurally unrelated E3 ubiquitin ligases, Neuralized (Neurl) and Mindbomb (Mib), mediate mono-ubiquitination of Notch ligands, which is necessary for ligand endocytosis. In mammals, there are two different Mindbomb paralogs (Mib1 and Mib2) and two Neuralized paralogs (Neurl1 and Neurl2). Analysis of germline and conditional-knockout mice of each Mib and Neurl gene has revealed that only deletion of Mib1 produces widespread Notch loss-of-function phenotypes. In contrast, loss of Mib2 or the two Neurl paralogs in mice is well tolerated and do not produce such phenotypes, indicating that Mib1 has an essential, nonredundant role in Notch ligand ubiquitination and trafficking in mammals. Similarly, in humans, single-allele familial mutations of Mib1 are correlated with left ventricular noncompaction cardiomyopathy (LVNC) and aberrant Notch signaling. However, it should be noted that Notch is not the only target of Mib1 and it is likely that Mib1 can interact with additional protein substrates. Together, these results suggest that ubiquitination and/or endocytosis is required for ligand competency. Additionally, ligand endocytosis after receptor binding has been implicated in generating the molecular force needed to confer conformational changes within the Notch receptor ectodomain that enables Adam-mediated cleavage. However, it is currently unclear how these disparate activities associated with Notch ligand endocytosis and trafficking enhance Notch signaling activity.
Productive Notch signaling is also dependent on Notch receptor endocytosis, which controls the rate of Notch degradation and, therefore, regulates the levels of functional Notch receptors at the cell surface. Several E3 ligases, including the Itchy/Nedd4 (neural precursor expressed, developmentally downregulated 4) family, act as negative regulators of Notch signaling that target nonactivated Notch for lysosomal trafficking and degradation. In many instances, loss of these ubiquitin ligases results in Notch overactivation, presumably due to the accumulation of Notch receptor within endosomal compartments where it becomes susceptible to ligand-independent proteolytic activation and NICD release. By contrast, a different E3 ligase, Deltex, promotes Notch signaling by antagonizing the negative effects of the Itchy/Nedd4 family and enhancing Notch accumulation in an endosomal compartment that promotes receptor recycling and ligand-independent activation. Unlike the essential role of Adam10 in ligand-dependent Notch S2 cleavage, Adam17 and possibly other proteases are responsible for ligand-independent Notch S2 cleavage in these specific endosomal compartments. Intriguingly, both murine Notch1 and Notch2, and human Notch1, require Adam17 for ligand-independent activation, whereas human Notch2 is resistant to such processing. While Notch1 NRR activating mutations that result in ligand-independent proteolysis are found frequently in human leukemias, highlighting the importance of tight control of metalloprotease access to the S2 site, it is currently unclear whether such ligand-independent Notch activation plays a role in mammalian development and normal tissue homeostasis. Recently, several Notch interacting proteins have been identified such as Drosophila Crumbs and mammalian Commd9 that regulate Notch localization and endosomal trafficking that impact ligand-independent activation. Together, these studies suggest that the balance of effects generated by different E3 ligases and other binding partners impacts the fate, sorting and compartment-specific degradation of Notch that ultimately control the cell surface half-life of Notch, its accessibility to functional ligand interactions as well as its ability to be processed by γ-secretases.
Numb is another well-characterized Notch antagonist in which asymmetric partitioning and loss of Notch activity occur in one daughter cell during mitosis. The mechanism by which Numb inhibits Notch activity is less well understood. Interestingly, studies in mammals have demonstrated that a Golgi protein (ACBD3) associates with Numb upon Golgi fragmentation during cell division, providing a mechanism for coupling asymmetric Notch signaling to mitosis. A recent study reported that the tumor suppressor miR-34a and Numb synergize to regulate asymmetric division and suppress plasticity in colon cancer stem cells. Similar processes of asymmetric segregation of Notch activity have been associated with Notch ligands and the asymmetric segregation of Neuralized, which promotes ligand endocytosis and activity.
Other E3 ubiquitin ligases, including Fbxw7, can act as tumor suppressors and have been implicated in the degradation of several proto-oncogenes, including NICD. CDK8 kinase-dependent phosphorylation of the NICD PEST domain makes it available for Fbxw7 ubiquitination and subsequent proteosomal degradation. Intestine-specific deletion of Fbxw7 resulted in impaired goblet cell differentiation and accumulation of proliferating progenitor cells and this was associated with increased Notch activity and c-Jun expression. Loss of Fbxw7 also accelerated Apc-dependent tumorigenesis confirming its role as a tumor suppressor in the intestine. Recently, Behrens and colleagues have proposed that Fbxw7 not only reduces the stability of NICD but also that the Fbxw7 gene is also transcriptionally downregulated by the Notch effector Hes5, creating a potential intracellular positive feedback loop for Notch signaling.
Lastly, more recent studies have shown that deubiquitination of Notch by ubiquitin-specific proteases Usp28 and Usp12 regulate Notch trafficking and activity. In colorectal cancer, Usp28 has been shown to reduce FBW7-dependent ubiquitin-mediated proteasomal degradation of NICD, thereby increasing Notch activity. Conversely, in vitro studies have demonstrated that upon Itch-dependent polyubiquitination of inactive uncleaved Notch, Usp12 deubiquitinates Notch and promotes its lysosomal degradation. Whether additional ubiquitin ligases and ubiquitin-specific proteases can regulate other core components of Notch signaling remain to be determined.
Notch signaling is an evolutionarily conserved mechanism for establishing physiological differences in neighboring cells ( Fig. 6.1 ). As described above, the signal is short lived due to the proteolytic cleavage of the receptor upon ligand activation and the subsequent rapid destruction of the released NICD signaling fragment. After ligand-receptor interaction, the key rate-limiting step for the initiation of a Notch signaling event is Adam10-mediated S2 cleavage; however, as outlined above, other types of regulation can occur at many steps of the pathway that modulate the activity of both ligands and receptors. These regulatory mechanisms may affect either the amplitude or duration of a signaling event. Evidence suggests that pathway activity is regulated by restriction of ligand or receptor availability (both spatially and temporally), modulation of ligand and receptor trafficking, and posttranslational modification of receptor and ligand. In addition to protein regulation, microRNAs are also involved in controlling pathway component availability. For example, in intestinal stem cells (ISCs) of the Drosophila gut, miR-305 regulates Notch and insulin pathways required for adaptive homeostasis associated with nutritional status. Another example is the tumor suppressor miR34a that binds to the 3′ UTR mRNA sequences of Notch receptors, leading to reduced Notch protein levels and decreased downstream Notch signaling. In colon cancer stem cells, miR-34a controls a bimodal Notch signal that specifies the choice between self-renewal and differentiation. More recent work demonstrated that miR-34a acts together with Numb and Notch to form a feed forward loop that curbs excessive proliferation by enforcing a binary cell fate choice and suppressing cancer stem cell plasticity. This field of research is still relatively nascent, and it is likely that mammalian Notch activity is differentially regulated based on temporal, tissue-specific, and cell-specific contexts.
The intestine is composed of cells originating from each of the three germ layers: the epithelium from endoderm, the mesenchyme (muscle, myofibroblasts, etc.) from mesoderm, and the enteric nervous system from ectoderm. The homeobox transcription factor caudal type homeobox 2 (Cdx2) appears to be a master regulator of intestinal identity, and its expression is essential for specification of the intestinal epithelium from the primordial gut endoderm, and for establishing normal epithelial-mesenchymal interactions. After morphogenesis of the intestine, stem and progenitor cells continuously divide and differentiate to maintain the epithelium throughout the lifespan of the organism. At least six distinct epithelial cell types are formed. This includes absorptive enterocytes and three secretory (granulocytic) cell types: mucus-producing goblet cells, antimicrobial peptide-producing Paneth cells, and hormone-releasing endocrine cells. Less frequent intestinal cell types include tuft cells, also called brush cells, which are chemosensory cells that orchestrate intestinal responses to parasite infection (also see Chapter 32 ), and microfold, or M, cells, which transport luminal antigens across the epithelium to mucosa-associated immune cells. The general structures of developing and adult intestine, including epithelial and mesenchymal components, are shown in Fig. 6.2 .
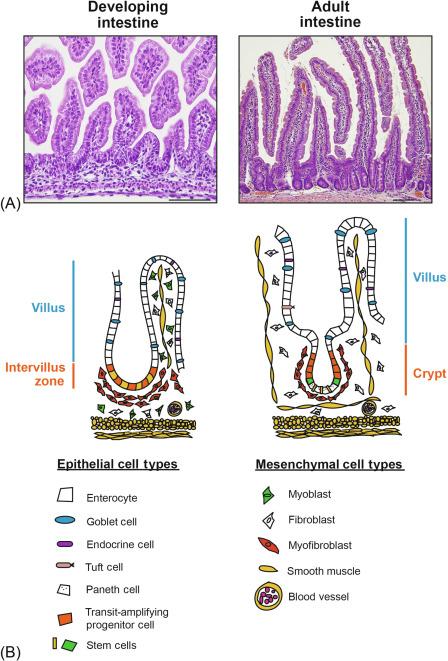
A complex network of signaling pathways and transcription factors work in concert to maintain homeostasis by regulating proliferation and cellular differentiation. Many studies have demonstrated the central importance of Notch signaling for homeostatic control of the intestinal epithelium, regulating both progenitor cell proliferation and cell fate determination. In addition, other fundamental signaling pathways are critical for intestinal development and homeostasis, including Wnt, Hedgehog, and bone morphogenetic protein (BMP). The specific roles of each of these pathways have been established primarily from analysis of genetically engineered mouse models. Before detailing the important functions of Notch signaling in the intestine, these other fundamental signaling pathways will be briefly summarized because they are somewhat interdependent and regulate overlapping functions.
In the intestine, Hedgehog signaling is exclusively paracrine. The pathway ligands Sonic Hedgehog (Shh) and Indian Hedgehog (Ihh) are secreted from epithelial cells and activate downstream signaling through their receptors Patched 1 and 2 and effectors Gli1, 2, and 3 in the mesenchyme. Gli2 appears to be the main effector for Hedgehog signaling in the developing intestine. At embryonic day 8.5 of mouse development (E8.5), Ihh and Shh are expressed in the gut endoderm in overlapping patterns, and by late fetal development, this expression pattern is restricted toward the proliferating epithelium of the intervillus zone. Mesenchymal cell clusters that form just beneath the epithelium at these zones are Hedgehog ligand responsive and drive villus formation starting at E14.5. Mice deficient in Shh or Ihh die perinatally and exhibit many gastrointestinal defects, including changes in enteric nervous system development, loss of smooth muscle, and altered epithelial proliferation. Similar phenotypes were observed in other studies that examined the consequences of blocking all Hedgehog signaling in the intestine by treating neonatal mice with a Hedgehog neutralizing antibody or a pharmacological inhibitor, or by using a genetic mouse model that expressed a secreted form of the pan-Hedgehog inhibitor Hhip from the intestinal epithelium via the villin promoter (Villin-Hhip mice). Phenotypes include increased epithelial proliferation and formation of ectopic crypt-like structures, impaired cluster formation and villus emergence, and reduced smooth muscle. Hedgehog signaling is also important for maintaining intestinal homeostasis in the adult. Mice with Cre-activated Hhip expression (VFHhip mice), or a conditional deletion of Ihh (Cyp1a1-CreIhh fl/fl ) in the intestinal epithelium showed expansion of the proliferative zone and progressive loss of smooth muscle.
In addition to its role in restricting proliferation and maintaining stromal differentiation, Hedgehog signaling has also been implicated in the inhibition of ISC self-renewal and differentiation, likely through suppressing Wnt signaling. Thus, the combined effect of Hedgehog signaling is important for villus formation, smooth muscle proliferation and differentiation, and restriction of the epithelial proliferative compartment in perinatal and adult intestine. Furthermore, since Hedgehog signaling mutants displayed epithelial phenotypes, a feedback mechanism that can signal from the mesenchyme back to the epithelium in response to Hedgehog signaling must exist.
Active BMP signaling is transduced through the cytoplasm to the nucleus by the SMAD transcription factors following BMP ligand-mediated receptor activation. BMP signaling in the intestine is bidirectional with multiple BMP ligands, receptors, SMAD transcription factors, and BMP inhibitors being expressed in both the epithelium and mesenchyme. Mesenchymal cells at intervillus and intercryptal regions have been shown to secrete BMP2 and BMP4 ligands. In the epithelium, it appears that the most active BMP signaling, as determined by the presence of nuclear phospho-SMAD1, 5, and 8 transcription factors, occurs in the villus rather than the crypts. This is likely due to inhibition of BMP signaling in the crypt region resulting from enriched expression of BMP inhibitors, including Noggin and Gremlin 1, in subcryptal mesenchymal cells. The pattern of BMP ligand and inhibitor expression leads to the formation of an increasing BMP activity gradient along the crypt-villus axis.
Phenotypic differences observed from modulating BMP signaling exclusively in the epithelium or mesenchyme suggests the pathway may play different roles in these two cellular compartments. A transgenic mouse in which the villin promoter was used to drive expression of Xenopus noggin in the intestinal epithelium (X-noggin), presented with ectopic crypt structures in 3-month-old mice and adenomatous foci development in older mice, suggesting that BMP signaling normally functions to limit crypt number. These types of epithelial changes are reminiscent of a rare, autosomal-dominant gastrointestinal syndrome called familial juvenile polyposis (FJP), which is characterized by development of hamartomatous polyps throughout the gut. Accordingly, mutations in SMAD4 and Bmpr1a have been identified in FJP patients, consistent with BMP signaling alterations in the pathogenesis of this syndrome. Indeed, a second mouse model in which the Bmpr1a receptor was conditionally deleted using the interferon-inducible Mx1-Cre also exhibited polyp formation. In contrast, loss of Bmpr1a in the epithelium only showed increased proliferation and a defect in secretory cell differentiation, but not formation of ectopic crypts or polyps, suggesting that epithelial BMP signaling is not sufficient for driving FJP. In agreement, depleting BMP signaling exclusively in pericryptal fibroblasts resulted in increased mesenchymal proliferation, development of a reactive stroma, and increased polyposis formation at 1 year. These reports suggest that it is mesenchymal BMP signaling that is primarily responsible for suppressing epithelial hyperproliferation.
It is thought that epithelial BMP signaling limits epithelial stem cell proliferation via restriction of ISC self-renewal. There is some evidence that BMP represses ISC self-renewal via Wnt pathway suppression ; however, this is uncertain as some studies showed no change in Wnt signaling after repression of epithelial BMP signaling, or observed that BMP regulation was Wnt-independent.
It is likely that BMP signaling is a key factor mediating Hedgehog effects on epithelial proliferation and villus morphogenesis. BMP4 and BMP7 are positively regulated by Hedgehog signaling, and the ectopic crypt phenotypes in the Hedgehog loss-of-function Villin-Hhip mouse and the BMP loss-of-function X-noggin mouse are similar. During development, Hedgehog-responsive mesenchymal clusters express BMP ligands and signal modifiers. Manipulating BMP signaling via BMP antagonist or exogenous BMP ligand administration, and conditional Bmpr1a deletion in Hedgehog-responsive mesenchyme, resulted in altered cluster formation, and diminished villus size and emergence. Together these studies demonstrate an important role for BMP in regulating intestinal development and ISC homeostasis.
In opposition to the antiproliferative effects of the Hedgehog and BMP signaling pathways, canonical Wnt signaling is a key pathway promoting proliferation in the intestinal crypts. Overactive Wnt signaling, such as that seen in the Apc min mouse model, leads to enlarged crypts with enhanced proliferation and progression to adenocarcinoma. Conversely, blocking Wnt signaling by forced expression of the secreted Wnt inhibitor Dickopff (Dkk1), or inactivation of the critical Wnt pathway components T-cell-specific transcription factor 4 (Tcf4), the signaling effector β-catenin, the Tcf4 target gene Myc , or deletion of R-spondin/Lgr complex components, leads to decreased proliferation and crypt loss. There is also evidence that noncanonical Wnt signaling through Wnt5a is important for aspects of gastrointestinal development, regeneration, and inflammation ; however, canonical Wnt signaling through β-catenin and Tcf4 appears to be most important for maintaining survival and promoting proliferation of ISCs. Accordingly, downstream targets of Tcf4-mediated Wnt signaling include proproliferation genes such as c-Myc and Cyclin D1.
Numerous Wnt ligands, receptors, and coreceptors are expressed in the intestine, with ligands produced in different epithelial cell populations as well as in pericryptal mesenchymal cells. The key cellular source and identification of specific Wnts functioning as ISC niche factors have been an area of active investigation. Global ablation of Wnt ligand secretion by pharmacologic inhibition of Porcupine (Porcn) led to reduced ISC numbers and blocked proliferation and regeneration. Surprisingly, ablation of Wnt ligand secretion in the epithelium via genetic deletion of Porcn did not disrupt intestinal homeostasis and regeneration, demonstrating that epithelial cells such as Paneth cells are not the key niche cell source of Wnts. Moreover, Porcn deletion in Myh11-positive subepithelial myofibroblasts did not affect intestinal homeostasis and regeneration. However, pericryptal Myh11-negative Foxl1-positive stromal cells have been shown to express Wnt family genes and are suggested to be a critical source of Wnt signaling by their secretion of Wnt activity potentiators, including Wnt 2b, Wnt5a, R-spondin 3, and the BMP inhibitors gremlin 1 and 2. Importantly, unlike Paneth cells, these mesenchymal cells were shown to be essential to support ISC proliferation. In addition to regulation of stem/progenitor cell proliferation, Wnt plays a role in cell fate determination in the intestine, with Wnt target genes Sox9 and EphB2/EphB3 mediating the differentiation of Paneth cells as well as their retention in the crypt base. From these studies, a simplified model of epithelial cell maintenance in the intestine includes stimulation of stem/progenitor cell proliferation by high Wnt in the crypt base, with cell cycle exit and terminal differentiation as cells move out of the crypt and the Wnt signaling zone.
All of the Notch ligands (Dll1, Dll3, Dll4, Jag1, and Jag2) and receptors (Notch1, 2, 3, 4) are expressed in the mouse gut during early development (E13.5) through adulthood with the exception of Dll3, which recedes after early development. Based on mRNA expression patterns, the Notch ligands Dll1, Dll4, and/or Jag1 and receptors Notch1 and Notch2 were thought to be the most likely mediators of epithelial Notch signaling in the adult intestine. It has not been fully determined which intestinal cell populations express specific ligands and receptors. It is likely that different cellular targets are involved with distinct aspects of Notch regulation, including stem cell maintenance, progenitor cell proliferation, cell fate specification, and possibly cell maturation. Identification of signal-receiving cells by immunostaining for NICD or Hes1 shows positive labeling in several epithelial cells in the crypts, suggesting that Notch signaling is primarily active in stem and progenitor cells. However, other than crypt base columnar (CBC) stem cells, the cellular identity of NICD- and Hes1-expressing cells have not yet been definitively established. Although Hes1 mRNA appears to be predominantly localized to the crypts, two reports have also shown nuclear Hes1 protein expression in villus enterocytes of the developing intestine. Finally, NICD protein was also observed in scattered goblet cells in one report. Thus, Notch signaling may also be active in some mature cells in the villus. Although the focus of this chapter is on epithelial Notch signaling, it is worthwhile to note that Notch signaling components are also expressed in the intestinal mesenchyme, where they are important for the development of the enteric nervous, vasculature, and lymphatic systems, and likely play a role in inflammatory cell function in the gut.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here