Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
lncRNAs promote tumorigenesis, chemotherapeutic resistance, and metastasis in bone sarcomas. Notable examples include MALAT1, TUG1, XIST, NEAT1, SNHG12, and UCA1.
lncRNAs have emerged as promising diagnostic and prognostic biomarkers, as well as veritable therapeutic targets in bone sarcomas.
Epigenetics encompasses the molecular changes that affect gene expression without direct alteration of the primary DNA sequence. Examples include DNA methylation, chromatin modification, nucleosome positioning, and dysregulation of noncoding RNAs (ncRNAs) [ ]. Historically, the majority of genetic studies have focused on protein-coding RNAs even though ncRNAs make up 98% of all human transcripts [ ]. These ncRNAs, which can be nonfunctional, can also serve as important epigenetic regulators [ ]. These ncRNAs are classified into three structural groups: small noncoding RNAs (sncRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs). The sncRNAs are less than 200 nucleotides, and include subtypes such as microRNAs (miRNAs), PIWI-associated RNAs (piRNAs), and transcription initiation RNAs (tiRNAs) [ , ]. Unlike the more common linear RNA variants, cirRNAs are loops of RNA generated by back-splicing, whereby the 3′ and 5′ ends of a linear RNA strand are covalently connected, and thus form an uninterrupted RNA “circle” [ ]. By definition, lncRNAs are greater than 200 nucleotides in length and will be the focus herein. Interestingly, although they are noncoding, some lncRNAs undergo the posttranscriptional modifications such as splicing, polyadenylation, and capping attributed to coding mRNA processing. lncRNAs are relatively common as they account for over 68% of all ncRNAs, and serve diverse modulatory functions through chromatin modification, transcription, mRNA stabilization, or direct action with other RNA species [ ]. With the development of whole-genome sequencing technology, lncRNAs have gained notoriety for their roles in cancer [ ]. Among the various malignancies they promote, sarcomas are clearly implicated [ ].
Bone sarcomas are rare malignancies originating from mesenchymal cells that often present within long bones of the extremity, pelvis, or vertebra. In 2019 there were approximately 3500 patients in the United States newly diagnosed with primary bone sarcomas, representing less than 0.2% of all new cancers [ ]. Total deaths were estimated at 1660 cases per year, which is 0.3% of cancer-related mortality [ ]. Although they are in fact infrequent, they cause considerable mortality and morbidity for inflicted patients as available treatments often fail to address poorly localized variants. Osteosarcoma is the most common bone sarcoma, followed by Ewing's sarcoma, chondrosarcoma, and chordoma [ ], all of which will be addressed in this chapter. And while the standard treatment protocol for osteosarcoma and Ewing's sarcoma is quite aggressive, consisting of perioperative chemotherapy or radiation and extensive surgical resection, outcomes have not significantly improved for decades. According to the Surveillance, Epidemiology, and End Results (SEER) database, the 5-year overall survival rate for patients with bone sarcoma is 66.2%, and for those with metastatic disease the 5-year overall survival rate drops to 10%–30% [ ]. In patients with chondrosarcoma or chordoma, even worse outcomes are common as they are especially resistant to available chemotherapy and radiotherapy. In response, there has been an expansion of genetic and molecular biology works focused on novel targets in bone sarcoma therapy. In this chapter, we summarize the expression and function of bone sarcoma–related lncRNAs and their clinical applications.
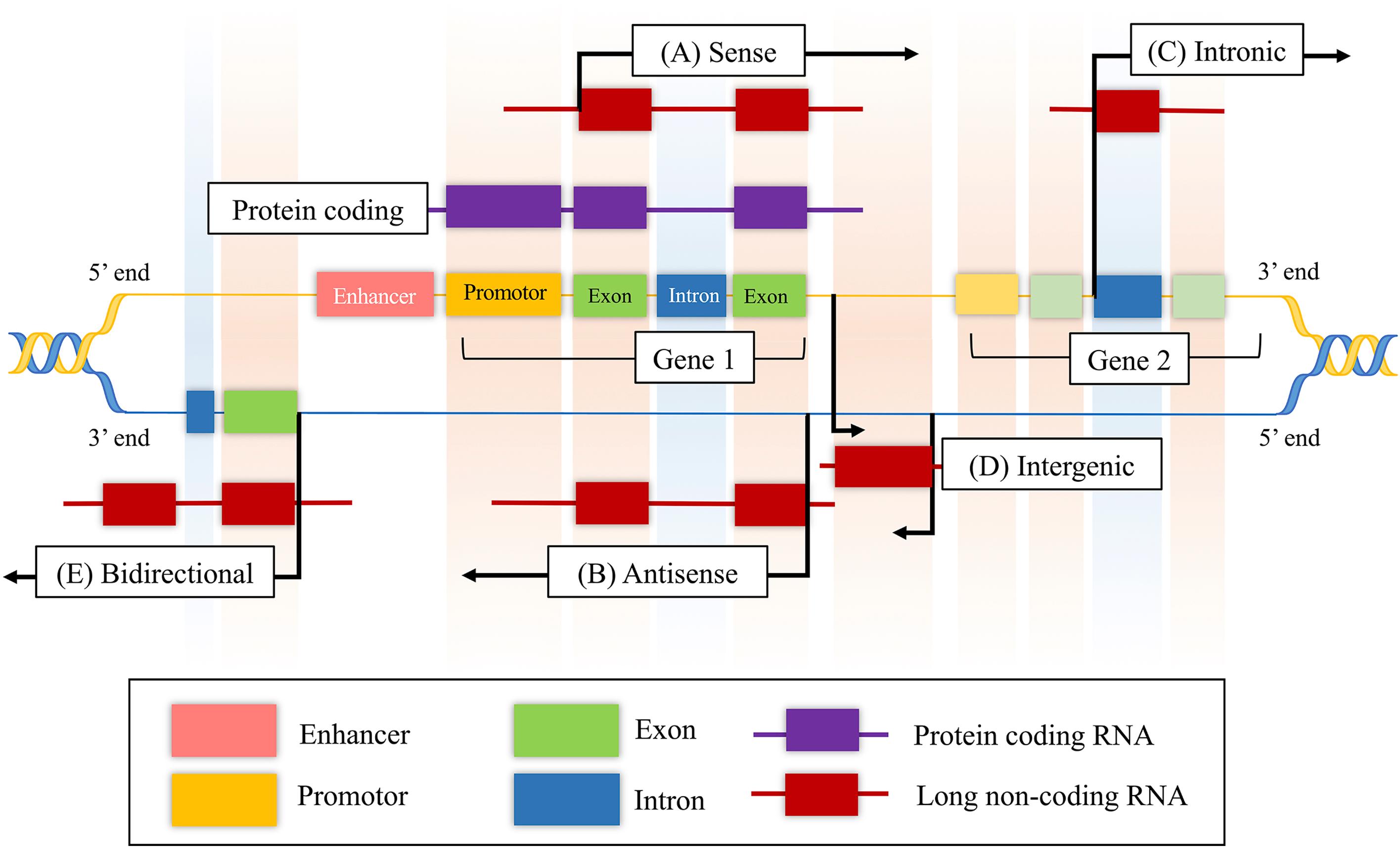
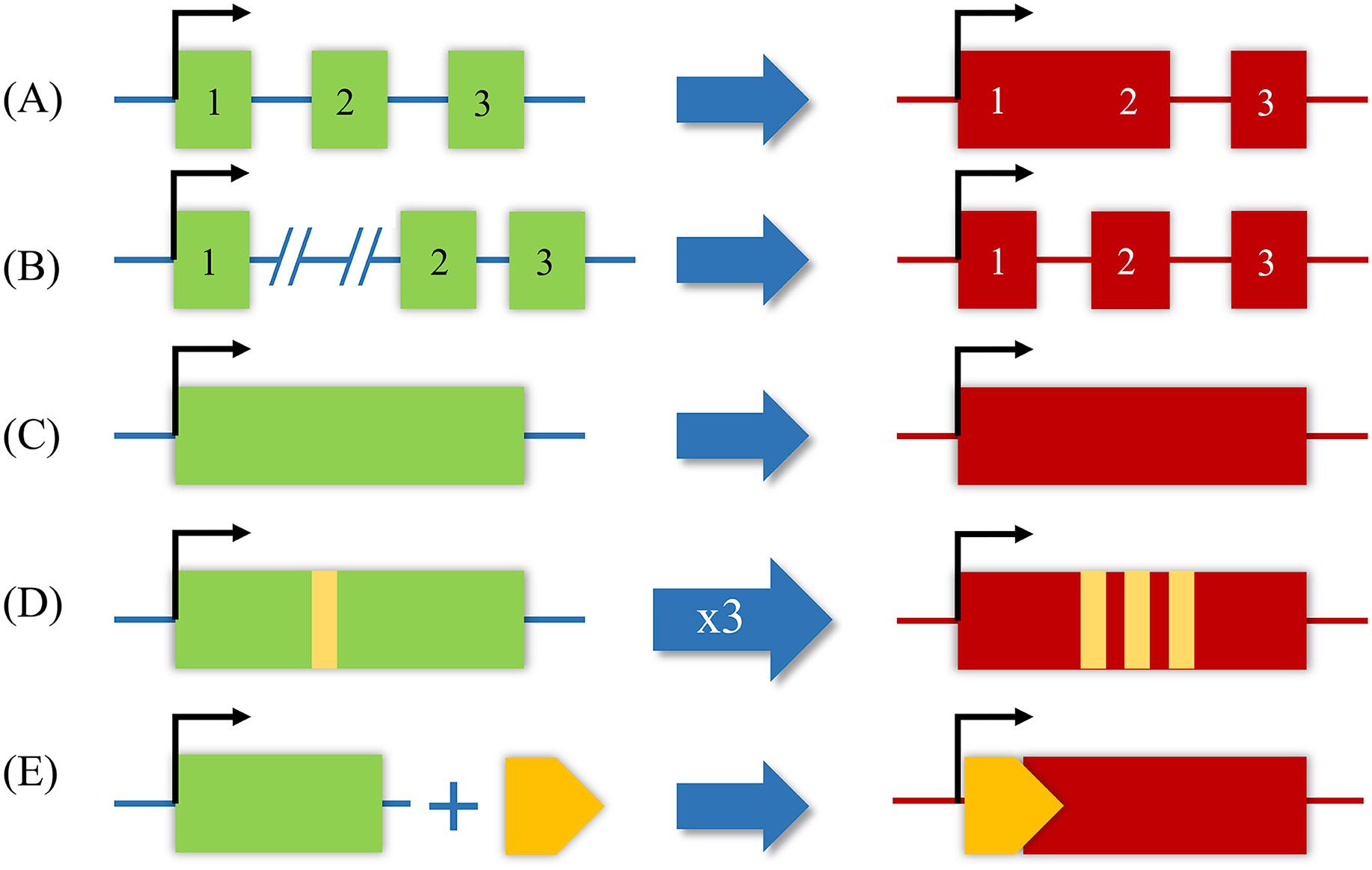
As shown in Fig. 34.1 , lncRNAs are classified according to their proximity to genomic protein-coding regions: (1) sense, (2) antisense, (3) intronic, (4) intergenic, and (5) bidirectional [ , , ]. Sense and antisense lncRNAs overlap at least one exon of another transcript on a consecutive strand [ , , ]. Intronic lncRNAs are transcribed from an intron without exon overlapping. Intergenic lncRNAs are transcribed from the genomic interval between two coding genes from either strand [ , , ]. Bidirectional lncRNAs are transcribed along with the complementary strand of a neighboring coding transcript in close genomic proximity [ , ]. As an additional measure of categorization, lncRNAs are known to have five possible origins: (1) a protein-coding gene is damaged and transformed into lncRNA, (2) a chromosomal rearrangement where two nontranscribed regions are juxtaposed and give rise to a multi-exon lncRNA, (3) duplication of a noncoding gene by retrotransposition creates either a functional gene or nonfunctional retropseudogene without protein-coding capacity, (4) two local, tandem duplication events generate neighboring repeats within noncoding RNA, and (5) insertion of a transposable sequence produces a functional noncoding RNA ( Fig. 34.2 ) [ ].
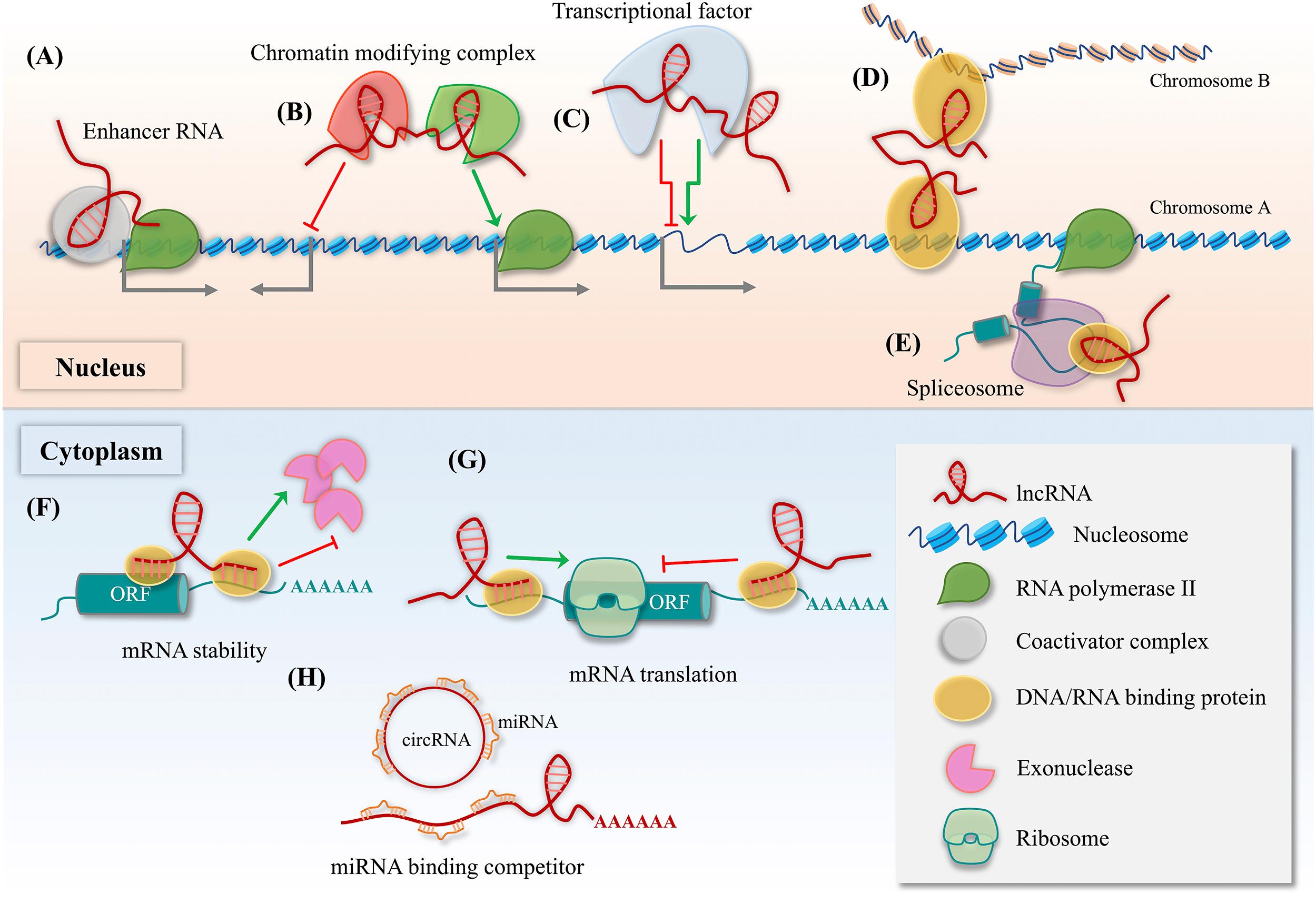
Whatever their origin, lncRNA may show abnormal expression in cancerous tissues and promote a deadly phenotype via several mechanisms. Within the nucleus, lncRNAs regulate transcription by acting as enhancer RNA, recruiting chromatin modifying complexes, or by integrating with transcriptional factors [ , ]. Alternatively, they can directly modify the spatial conformation of chromosomes or pre-mRNA splicing ( Fig. 34.3 ) [ , ]. Within the cytoplasm, lncRNAs regulate mRNA stability, translation, or compete with miRNA-binding sites [ ]. With direct protein binding, they affect DNA methylation, histone methylation, acetylation, ubiquitination, and organelle formation [ , ]. Emerging research has made clear that lncRNAs have robust oncogenic or tumor-suppression roles, and through their large-scale regulatory processes stimulate tumorigenesis, proliferation, metastasis, and chemotherapeutic resistance in cancers such as bone sarcomas [ , , ].
Despite wide surgical resection and aggressive perioperative chemotherapy, the 5-year overall survival rate has plateaued at 70% for past four decades. Highly anticipated cancer immunotherapy works, which have been efficacious in several other malignancies, have shown relatively poor results in osteosarcoma [ ]. Moreover, the tendency for osteosarcoma to undergo pulmonary metastasis, recur, and develop chemotherapeutic resistance remains significant obstacles to therapy [ ]. Given these barriers, there has been an emergence of proposed novel targets and therapeutics that precisely target the aberrancies of osteosarcoma, including works in lncRNAs. In an lncRNA microarray analysis, 25,733 lncRNAs were compared between nine human primary osteosarcoma tissues and their paired adjacent normal tissue samples, and researchers found 403 lncRNAs upregulated and 798 downregulated [ ]. Furthermore, a pathway analysis concluded the differential expression was a result of known signaling pathways, with 34 pathways upregulated and 32 pathways downregulated [ ]. These findings and others have prompted an emerging and recent body of research outlining the crucial roles of lncRNAs in osteosarcoma growth, proliferation, metastasis, chemotherapeutic resistance, and impact on patient outcomes.
lncRNAs have consistently shown to promote osteosarcoma pathogenesis via several pathways. High expression of the nuclear lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is present in both osteosarcoma cell lines and tissues [ ] and promotes cell proliferation, invasion, migration, and metastasis by increasing c-Met and SOX4 expression [ ]. MALAT1 is also involved in miR-376A/transforming growth factor α (TGF-α) axis–mediated osteosarcoma proliferation and progression [ ]. In one study, MALAT1 promoted osteosarcoma antiapoptotic and proliferative activity in vitro and in vivo via upregulation of histone deacetylase 4 (HDAC4) and decoying of miR-140-5p [ ]. Along with the high-mobility group protein B1 (HMGB1), MALAT1 expression also correlated with osteosarcoma cell proliferation via the MALAT1/miR-142-3p/miR-129-5p/HMGB1 axis [ ]. In a xenograft model, MALAT1 inhibition delayed osteosarcoma growth via the Ras homolog family member A (RhoA)/Rho-associated coiled-coil containing protein kinases (ROCKs) pathway [ ]. In another murine model, lncRNA H19 was highly upregulated in both xenograft and human primary osteosarcoma tissues, and was induced by the hedgehog signaling pathway and yes-associated protein 1 (Yap1) [ ]. Inhibition of the hedgehog signaling pathway or Yap1 gene knockdown was able to downregulate H19, resulting in decreased osteosarcoma cell viability [ ]. In contrast, expression of BRAF-regulated lncRNA 1 (BANCR) was low in the MG-63 osteosarcoma cell line. When overexpressed, BANCR inhibited osteosarcoma proliferation and promoted apoptosis by upregulation of phosphorylated c-Jun N-terminal kinase (JNK) and downregulation of the Wnt/β-catenin pathway [ ].
One notable way lncRNAs cause osteosarcoma proliferation is by interacting with miRNAs. The lncRNA taurine upregulated gene 1 (TUG1), which is highly expressed in human osteosarcoma tissues and cell lines [ , ], can undergo successful siRNA knockdown and lead to decreased osteosarcoma proliferation and increased apoptosis [ ]. On such study showed suppression of TUG1 inhibits osteosarcoma cell proliferation and colony formation by inducing G0/G1 cell cycle arrest and apoptosis in vitro , and shows decreased in vivo tumor growth [ ]. Other works have revealed TUG1 competes with endogenous RNA (ceRNA) to inflect specific miRNA functions in osteosarcoma. For example, it can directly sponge miR-9-5p and therefore knockdown its function, which decreases POU class 2 homeobox 1 (POU2F1) expression and activity. Additionally, TUG1 promotes osteosarcoma tumorigenesis by upregulating enhancer of zeste homolog 2 (EZH2) via miR-144-3p and the Wnt/β-catenin pathway [ ]. Various other miRNAs are sponged by TUG1, and include miR-153, miR-212-3p, miR-132-2p/SOX4, miR-140-5p/profilin2, and miR-212-3p/FOXA1 [ ]. In a unique combined mechanism, TUG1 has also been shown to promote osteosarcoma proliferation through modulation of runt-related transcription factor 2 (RUNX2) [ ], or by affecting glycolysis via activity of hexokinase 2 (HK2) [ ].
Upregulation of lncRNA zinc finger E-box binding homeobox 1 antisense RNA 1 (ZEB1-AS1) has correlated with size, staging, metastasis, and shorter overall survival in osteosarcoma patients [ ]. Mechanistically, enhancement of ZEB1-AS1 promotes osteosarcoma proliferation and migration by direct binding and recruitment of p300 to the ZEB1 promoter region, inducing an open chromatin and activated ZEB1 transcription profile [ ].
Greater expression of lncRNA X-inactive-specific transcript (XIST) was significantly more expressed in osteosarcoma cell lines and tissues compared to adjacent normal bone tissues, and correlated with advanced stage, tumor size, and distant metastasis [ , ]. XIST was also an independent risk factor for overall survival of osteosarcoma patients as it promoted proliferation, invasion, and migration, while also inhibiting cell cycle arrest and apoptosis through several pathways [ ]. Mechanistically, XIST repressed p21 expression and bound with EZH2 to stimulate proliferation [ ]. In addition, XIST functioned as a ceRNA by competitively sponging miRNAs such as miR-375-3p/AKT/mTOR, miR-320b/Ras-related protein RAP2B, miR-193a-3p/RSF1, miR-21-5p/PDCD4, miR-195-5p/Yap, and miR-137 [ ]. In a xenograft model, XIST inhibition suppressed osteosarcoma growth and induced apoptosis via activation of the nuclear factor-kappa B (NF-kB)/p53 upregulated modulator of apoptosis (PUMA) pathway [ ].
Some lncRNAs promote cancer through activation of other known tumorigenic factors. The oncogenic tyrosine kinase protein C-Met, for example, correlates with MALAT1 and synergistically promotes osteosarcoma proliferation and metastasis [ ]. The lncRNA plasmacytoma variant translocation 1 (PVT1) functions by enhancing chemotherapeutic resistance of osteosarcoma through the c-Met/PI3K/AKT pathway [ ]. The positive feedback loop of the lncRNA HOXA transcript at the distal tip (HOTTIP) is able to increase expression of c-Myc, a prominent transcription factor in osteosarcoma, leading to invasion and migration [ ].
lncRNAs can also facilitate osteosarcoma proliferation by controlling the expression profile of several oncogenes. The lncRNA homeobox (HOX) transcript antisense intergenic RNA (HOTAIR), for example, is highly expressed in human osteosarcoma tissues with significant correlation to advanced stage, higher histological grade, and poorer prognosis [ ]. In an in vitro experiment, knockdown of HOTAIR suppressed osteosarcoma proliferation and invasion via downregulation of matrix metalloproteinase 2 (MMP-2), MMP-9, TGF-β, B-cell lymphoma 2 (Bcl-2), and upregulation of p53 and tumor necrosis factor α (TNF-α) [ , ]. Various other lncRNAs have been reported to affect osteosarcoma cell growth and proliferation, as summarized in Table 34.1 .
| lncRNA | Expression | Role of lncRNA | Potential mechanism | Reference |
|---|---|---|---|---|
| MALAT1 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis ↓ Apoptosis |
RhoA/ROCK pathway, c-MET and Sox4 expression, MALAT1/miR-376A/TGF-α axis, HDAC4/miR-140-5p axis, ceRNA binding to miR-34a/c-5p and miR-449a/b, MALAT1/miR-142-3p/miR-129-5p/HMGB1 axis | [ ] |
| TUG1 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis ↓ Apoptosis |
Modulating RUNX2 expression, modulating glycolysis via HK2 inhibit G0/G1 cell cycle arrest, as ceRNA sponging miRNAs such as miR-9-5p/POU2F1, miR-144-3p/EZH2/Wnt/β-catenin pathway, miR-153, miR-212-3p, miR-132-3p/SOX4, miR-140-5p/PFN2, miR-212-3p/FOXA1 | [ ] |
| XIST | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis ↓ Apoptosis |
Repression of p21, binding with EZH2, as ceRNA sponging miRNAs such as miR-375-3p/AKT/mTOR, miR-320b/Ras-related protein RAP2B, miR-193a-3p/RSF1, miR-21-5p/PDCD4, miR-195-5p/Yap, miR-137 activation of NF-kB/PUMA pathway | [ ] |
| H19 | ↑ | ↑ Growth and proliferation ↓ Apoptosis |
Hedgehog signaling pathway and oncogene Yap1, interaction with miR-141 | [ ] |
| ZEB-1AS1 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis |
Recruiting and binding p300 to the ZEB1 promotor region, leading to activation of ZEB1 transcription | [ ] |
| HOTAIR | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis |
Activation of MMP-2, MMP-9, TGF-β, Bcl-2, and downregulation of p53 and TNF-α | [ , ] |
| HNF1A-AS1 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis |
Wnt/β-catenin pathway | [ ] |
| HOTTIP | ↑ | ↑ Invasion, migration, metastasis | Positive feedback loop of HOTTIP/c-Myc | [ ] |
| PVT1 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis ↓ Apoptosis |
Upregulation of Bcl-2, cyclin D1, FASN expression via negatively regulating of miR-195, c-Met/PI3K/AKT pathway | [ , ] |
| MFI2 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration ↓ Apoptosis |
Upregulation of FOXP4 | [ ] |
| ANCR | ↑ | ↑ Growth and proliferation ↑ Invasion, migration ↓ Apoptosis, G0/G1 cell cycle arrest |
Upregulation of EZH2, E-cadherin, N-cadherin, and phosphorylated p38MAPK, downregulation of p21, p27 expression | [ ] |
| SNHG12 | ↑ | ↑ Growth and proliferation ↑ Invasion, migration, metastasis |
Upregulation of AMOT mRNA expression, modulating the expression of Notch2 and IGF1R by sponging miR-195-5p | [ , ] |
| UCA1 | ↑ | ↑ Growth and proliferation both in vitro and in vivo ↑ Invasion, migration, metastasis ↓ Apoptosis |
Inactivation of PTEN/AKT signaling pathway, regulation of miR-301a and CXCR4 expression, miR-182 and TIMP2, CREB-1-mediated PI3K/AKT/mTOR pathway, PI3K/AKT/GSK3β, and NF-κB pathways | [ ] |
| NEAT1 | ↑ | ↑ Growth and proliferation both in vitro and in vivo ↑ Invasion, migration, metastasis ↓ Apoptosis |
As ceRNA sponging miRNAs such as miR-186-5p, miR-339-5p, miR-194, and miR-34a-5p. Suppression of E-cadherin expression through the interaction with G9a–DNMT1–snail complex | [ ] |
| HIF2PUT | ↓ | ↓ Growth and proliferation ↓ Invasion, migration |
Controlling HIF-2α expression | [ ] |
| BANCR | ↓ | ↓ Growth and proliferation ↓ Invasion, migration ↑ Apoptosis |
JNK and Wnt/β-catenin pathway | [ ] |
| TUSC7 | ↓ | ↓ Proliferation and colony formation both in vitro and in vivo ↓ Invasion, migration ↑ Apoptosis |
Regulation by miR-211 | [ ] |
| MEG3 | ↓ | ↓ Growth and proliferation ↓ Invasion, migration, metastasis ↑ Apoptosis |
Regulation of p53 (positively) and MDM2 The MEG3/miR-361-5p/FoxM1 axis |
[ ] |
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here