Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Ligand binding and activation of cell surface and internal receptors trigger the activation and/or suppression of signaling cascades that regulate diverse cellular processes including cell growth, proliferation, survival, and invasion, among others.
Multiple nodes within these intracellular signaling networks are genetically and epigenetically altered in human cancers, leading to constitutive pathway activation or suppression.
Some cancers are dependent on genomic alterations in oncogenes or tumor suppressor genes for their growth and survival, a phenomenon known as oncogene addiction.
Drugs that selectively target mutated proteins critical for the maintenance of the transformed phenotype have shown unprecedented clinical activity in genetically defined cancer subsets.
Precision medicine refers to the use of genetic and epigenetic information unique to an individual cancer patient to develop treatment regimens that target the driver oncogenes and tumor suppressors responsible for tumor progression. Potential challenges to the application of this approach include the current inability to directly inhibit some oncogenic proteins (i.e., mutant KRAS), the development of drug resistance, technical hurdles posed by limited tissue availability for prospective molecular characterizing, and intratumoral and lesion-to-lesion genomic heterogeneity.
Routine genomic analysis of tumors or tumor-derived cell free DNA in plasma is now a component of standard care in an increasing number of cancer types, with the results used to guide treatment selection.
The underlying basis of the cancer phenotype is deregulated cell growth, which stems from two main hallmarks of cancer: uncontrolled proliferation, and loss of programmed cell death (enhanced survival). In normal cells, these processes are tightly controlled through integration of signaling cascades that translate extracellular and intracellular cues into specific output responses. These signaling pathways are often initiated on binding of ligand to the extracellular domain of a receptor, followed by recruitment of adaptor proteins or kinases that activate an intracellular cascading network of protein and lipid intermediaries that ultimately produce a cellular response. In normal cells, the specificity, amplitude, and duration of signaling are tightly regulated, and these constraints are often abrogated in human cancers.
Investigation of the signal transduction pathways that regulate normal cellular functions has revealed that key components of these networks are commonly altered in cancer cells by mutation, amplification and deletion, chromosomal translocation, overexpression, or epigenetic silencing. These alterations lead to activation or suppression of signaling cascades that underlie the various hallmarks of the cancer phenotype. This chapter reviews the major signal transduction cascades, with a focus on those that are frequently altered in human cancers. Individual sections highlight signaling intermediaries that have been validated as drug targets in patients with cancer. Table 2.1 summarizes actionable gene-level and mutation-level alterations in cancer and the drugs that are currently approved by the US Food and Drug Administration (FDA) for treatment, that are recommended standard of care biomarkers, or that have promising clinical or preclinical efficacy.
| Gene | Variant | Cancer Type | Drug | Evidence a |
|---|---|---|---|---|
| ABL1 | BCR-ABL1 fusion | ALL | Dasatinib | 1 |
| Imatinib | 1 | |||
| CML | Dasatinib | 1 | ||
| Imatinib | 1 | |||
| Nilotinib | 1 | |||
| AKT1 | E17K | Breast | AZD5363 | 3 |
| Ovarian | AZD5363 | 3 | ||
| All Tumors | ARQ 751 | 4 | ||
| ALK | Fusions | NSCLC | Alectinib | 1 |
| Ceritinib | 1 | |||
| Crizotinib | 1 | |||
| Oncogenic mutations | NSCLC | Brigatinib | 1 | |
| Fusions | Soft tissue sarcoma | Ceritinib | 2 | |
| Crizotinib | 2 | |||
| L1196M, L1196Q | NSCLC | Brigatinib | 3 | |
| R1275Q | Embryonal tumor | Crizotinib | 4 | |
| ARAF | S214A | Histiocytosis | Sorafenib | 3 |
| S214C | NSCLC | Sorafenib | 3 | |
| ATM | N2875K, R3008C, truncating mutations | Prostate cancer | Olaparib | 4 |
| BRAF | V600D, V600E, V600G, V600K, V600M, V600R | Melanoma | Cobimetinib + vemurafenib | 1 |
| Dabrafenib | 1 | |||
| Dabrafenib + trametinib | 1 | |||
| Vemurafenib | 1 | |||
| Histiocytosis | Vemurafenib | 2 | ||
| NSCLC | Dabrafenib | 2 | ||
| Dabrafenib + trametinib | 2 | |||
| Vemurafenib | 2 | |||
| Colorectal | Binimetinib + cetuximab + encorafenib | 3 | ||
| Panitumumab + vemurafenib | 3 | |||
| Colorectal | Fluorouracil + radiation + trametinib | 4 | ||
| V600E, V600K | Melanoma | Trametinib | 1 | |
| Fusions | Ovarian | Paclitaxel + selumetinib | 3 | |
| K601E | Melanoma | Trametinib | 3 | |
| L597Q, L597R, L597S, L597V | Melanoma | Trametinib | 3 | |
| D594E, D594N, G466V, G469A, G469V, G596C | Melanoma | Trametinib | 4 | |
| KIAA1549-BRAF Fusion | Soft tissue sarcoma | Sorafenib + temsirolimus | 4 | |
| L597Q, L597V | Melanoma | BGB659 | 4 | |
| BRCA1 | Oncogenic mutations | Ovarian | Niraparib | 1 |
| Rucaparib | 1 | |||
| Olaparib | 2 | |||
| BRCA2 | Oncogenic mutations | Ovarian | Niraparib | 1 |
| Rucaparib | 1 | |||
| Olaparib | 2 | |||
| CDK4 | Amplification | Dedifferentiated liposarcoma | Abemaciclib, palbociclib | 2 |
| Well-differentiated liposarcoma | Abemaciclib, palbociclib | 2 | ||
| CDKN2A | Oncogenic mutations | Breast | Letrozole + palbociclib | 4 |
| Esophagogastric | Palbociclib | 4 | ||
| EGFR | 729_761del, 729_761indel, L858R | NSCLC | Afatinib | 1 |
| Erlotinib | 1 | |||
| Gefitinib | 1 | |||
| Osimertinib | 4 | |||
| E709_T710delinsD, E709K, G719A, G719C, G719D, G719S, A763_Y764insFQEA, L747P, A750P, A763_Y764insFQEA, L833V, L861Q, L861R, S768I, EGFR-KDD | NSCLC | Afatinib, erlotinib, gefitinib | 1 | |
| T790M | NSCLC | Osimertinib | 1 | |
| 762_823ins | NSCLC | EGF816 | 4 | |
| 762_823ins, G719A, L861R, S768I | NSCLC | AP32788 | 4 | |
| ERBB2 | Amplification | Breast | Ado-trastuzumab emtansine | 1 |
| Lapatinib | 1 | |||
| Lapatinib + trastuzumab | 1 | |||
| Pertuzumab + trastuzumab | 1 | |||
| Trastuzumab | 1 | |||
| Esophagogastric | Trastuzumab | 1 | ||
| Breast | Neratinib | 3 | ||
| Oncogenic mutations | Breast | Neratinib | 3 | |
| V659E | NSCLC | Lapatinib | 3 | |
| E770_K831indel, E770_K831ins | NSCLC | AP32788 | 4 | |
| ERCC2 | Oncogenic mutations | Bladder | Cisplatin | 3 |
| ESR1 | Oncogenic mutations | Breast | AZD9496, fulvestrant | 3 |
| D538G, Y537S | Breast | GDC-0810 | 4 | |
| EZH2 | Oncogenic mutations | Diffuse large B-cell lymphoma | GSK126 | 4 |
| Tazemetostat | 4 | |||
| FGFR1 | Amplification | Lung squamous cell carcinoma | AZD4547 | 3 |
| Debio1347 | 3 | |||
| BCR-FGFR1 fusion | Leukemia | Ponatinib | 4 | |
| FGFR2 | Fusions | Adrenocortical carcinoma | Debio1347 | 3 |
| JNJ-42756493 | 3 | |||
| Bladder | Debio1347 | 3 | ||
| JNJ-42756493 | 3 | |||
| Cholangiocarcinoma | BGJ398 | 3 | ||
| Debio1347 | 3 | |||
| Endometrial | Debio1347 | 3 | ||
| JNJ-42756493 | 3 | |||
| FGFR3 | Fusions | Adrenocortical carcinoma | Debio1347 | 3 |
| JNJ-42756493 | 3 | |||
| Bladder | Debio1347 | 3 | ||
| JNJ-42756493 | 3 | |||
| Glioma | Debio1347 | 3 | ||
| JNJ-42756493 | 3 | |||
| G370C, G380R, K650E, K650M, K650N, K650Q, K650R, K650T, R248C, S249C, S371C, Y373C | Bladder | Debio1347 | 3 | |
| JNJ-42756493 | 3 | |||
| Breast | Debio1347 | 4 | ||
| FLT3 | Y572_Y630ins | AML | Sorafenib | 3 |
| IDH1 | R132C, R132G, R132H, R132Q, R132S | AML | AG-120 | 3 |
| All tumors | BAY1436032 | 4 | ||
| CB-839 | 4 | |||
| IDH2 | R140Q, R172G, R172K, R172M, R172S | All liquid tumors | AG-221 | 3 |
| JAK2 | PCM1-JAK2 fusion | Leukemia | Ruxolitinib | 3 |
| KIT | 449_514mut, 550_592mut, A502_Y503dup, D579del, D820G, E554_K558del, H697Y, K550_W557del, K558delinsNP, L576P, P551_M552del, V555_L576del, V560D, V560del, V654A | GIST | Sunitinib | 1 |
| Thymic tumor | Sunitinib | 2 | ||
| 449_514mut, 550_592mut, D419del, D579del, E554_I571del, E554_K558del, E554_V559del, F522C, I563_L576del, I653T, K550_W557del, K558N, K558_E562del, K558_V559del, K558delinsNP, K642E, L576P, M541L, M552_W557del, N564_Y578del, N822H, N822Y, P573_D579del, P577_W582delinsPYD, P838L, Q556_K558del, T417_D419delinsI, T417_D419delinsRG, T574insTQLPYD, V530I, V555_L576del, V555_V559del, V559C, V559D, V559G, V559_V560del, V559del, V560D, V560G, V560del, V569_L576del, W557G, W557R, W557_K558del, Y553N, Y553_K558del, Y570H, Y578C | GIST | Imatinib | 1 | |
| 449_514mut, 550_592mut, D820G, D820Y, K550_W557del, K558delinsNP, N822K, V560D | GIST | Regorafenib | 1 | |
| D816F, D816Y, D820G, D820Y, L576P, N822I, V559D, V560G, W557_K558del | GIST | Dasatinib | 2 | |
| D816V, D820A, D820G, D820Y, K642E, L576P, V555_L576del, V559C, V559D, V654A, W557_K558del | GIST | Nilotinib | 2 | |
| D820A, D820E, D820G, D820Y, K642E, N505I, P577_D579del, V559D, W557_K558del | GIST | Sorafenib | 2 | |
| Thymic tumor | Sorafenib | 2 | ||
| K642E, L576P, V559A | Melanoma | Imatinib | 2 | |
| KRAS | Wild type | Colorectal | Cetuximab | 1 |
| Panitumumab | 1 | |||
| Regorafenib | 1 | |||
| Cabozantinib + panitumumab | 4 | |||
| Panitumumab + regorafenib | 4 | |||
| Pembrolizumab | 4 | |||
| Oncogenic mutations | All tumors | alpelisib + binimetinib | 4 | |
| Cobimetinib + GDC-0994 | 4 | |||
| Colorectal | Atezolizumab + cobimetinib | 4 | ||
| Fluorouracil + radiation therapy + trametinib | 4 | |||
| NSCLC | Abemaciclib, PD0325901 + palbociclib, palbociclib, ribociclib, ribociclib + trametinib | 4 | ||
| Binimetinib + erlotinib | 4 | |||
| Binimetinib, selumetinib, trametinib | 4 | |||
| Docetaxel + trametinib | 4 | |||
| MAP2K1 | Oncogenic mutations | Histiocytic disorder | Cobimetinib, selumetinib, trametinib | 3 |
| Low-grade serous ovarian | Cobimetinib, selumetinib, trametinib | 3 | ||
| Melanoma | Cobimetinib, selumetinib, trametinib | 3 | ||
| NSCLC | Cobimetinib, selumetinib, trametinib | 3 | ||
| MDM2 | Amplification | Liposarcoma | DS-3032b | 3 |
| RG7112 | 3 | |||
| SAR405838 | 4 | |||
| MET | 963_D1010splice, 981_1028splice, X1006_splice, X1007_splice, X1008_splice, X1009_splice, X1010_splice, X963_splice | NSCLC | Crizotinib | 2 |
| Cabozantinib | 3 | |||
| Capmatinib | 3 | |||
| Amplification | NSCLC | Crizotinib | 2 | |
| RCC | Cabozantinib | 2 | ||
| D1010H, D1010N, D1010Y | NSCLC | Crizotinib | 2 | |
| Cabozatinib | 3 | |||
| Capmatinib | 3 | |||
| MTOR | E2014K | Bladder | Everolimus | 3 |
| C1483F, F1888L, L2230V, S2215F, T1977K | RCC (clear cell) | Everolimus, rapamycin, temsirolimus | 4 | |
| NF1 | Oncogenic mutations | Glioblastoma | Trametinib | 4 |
| Melanoma | Trametinib | 4 | ||
| Neurofibroma | Binimetinib | 4 | ||
| Neurofibroma | PLX3397 | 4 | ||
| NRAS | Oncogenic mutations | Colorectal | Atezolizumab + cobimetinib | 3 |
| Melanoma | Binimetinib | 3 | ||
| Binimetinib + ribociclib | 3 | |||
| Thyroid | Radioiodine uptake therapy + selumetinib | 3 | ||
| Colorectal | Fluorouracil + radiation therapy + trametinib | 4 | ||
| NTRK1 | Fusions | All tumors | Larotrectinib | 3 |
| Salivary gland | Entrectinib | 3 | ||
| NTRK2 | Fusions | Salivary gland | Entrectinib | 3 |
| Larotrectinib | 3 | |||
| NTRK3 | Fusions | Salivary gland | Entrectinib | 3 |
| Larotrectinib | 3 | |||
| PDGFRA | FIP1L1-PDGFRA fusion | Leukemia | Imatinib | 1 |
| Fusions | Myelodysplasia | Imatinib | 1 | |
| Myeloproliferative Neoplasm | Imatinib | 1 | ||
| 560_561insER, A633T, C450_K451insMIEWMI, C456_N468del, C456_R481del, D568N, D842I, D842_H845del, D842_M844del, D846Y, E311_K312del, G853D, H650Q, H845Y, H845_N848delinsP, I843del, N659K, N659R, N659S, N848K, P577S, Q579R, R748G, R841K, S566_E571delinsR, S584L, V469A, V536E, V544_L545insAVLVLLVIVIISLI, V561A, V561D, V561_I562insER, V658A, W559_R560del, Y375_K455del, Y555C, Y849C, Y849S | GIST | Imatinib | 2 | |
| D842V | GIST | Dasatinib | 2 | |
| PDGFRB | Fusions | Dermatofibrosarcoma Protuberans | Imatinib | 1 |
| Myelodysplasia | Imatinib | 1 | ||
| Myeloproliferative neoplasm | Imatinib | 1 | ||
| PIK3CA | Oncogenic mutations | Breast | Alpelisib | 3 |
| Alpelisib + fulvestrant | 3 | |||
| Buparlisib | 3 | |||
| Buparlisib + fulvestrant | 3 | |||
| Copanlisib | 3 | |||
| Fulvestrant + taselisib | 3 | |||
| GDC-0077 | 3 | |||
| Serabelisib | 3 | |||
| Taselisib | 3 | |||
| Alpelisib + everolimus | 4 | |||
| Alpelisib + letrozole, Alpelisib + letrozole + ribociclib | 4 | |||
| Alpelisib + LJM716 + trastuzumab | 4 | |||
| Alpelisib + olaparib, buparlisib + olaparib | 4 | |||
| AZD5363 + fulvestrant | 4 | |||
| AZD8835 + fulvestrant | 4 | |||
| MLN0128 + serabelisib | 4 | |||
| All tumors | ARQ 751 | 4 | ||
| AZD5363 + olaparib | 4 | |||
| GDC-0077 | 4 | |||
| Endometrial | Alpelisib + fulvestrant | 4 | ||
| Buparlisib + fulvestrant | 4 | |||
| Fulvestrant + taselisib | 4 | |||
| Ovarian | Alpelisib + fulvestrant | 4 | ||
| Buparlisib + fulvestrant | 4 | |||
| Fulvestrant + taselisib | 4 | |||
| PTCH1 | Truncating mutations | Embryonal tumor | Sonidegib | 3 |
| Skin cancer, nonmelanoma | Sonidegib | 3 | ||
| Vismodegib | 3 | |||
| PTEN | Oncogenic mutations | All tumors | ARQ 751 | 4 |
| AZD5363 + olaparib | 4 | |||
| AZD8186 | 4 | |||
| Gedatolisib + palbociclib | 4 | |||
| GSK2636771 | 4 | |||
| LY3023414 | 4 | |||
| Endometrial | Olaparib | 4 | ||
| Prostate | Enzalutamide + LY3023414 | 4 | ||
| RAF1 | S257L | Lung adenocarcinoma | Sorafenib | 4 |
| RET | Fusions | NSCLC | Cabozantinib | 2 |
| Vandetanib | 3 | |||
| ROS1 | Fusions | NSCLC | Crizotinib | 1 |
| D2033N | Cabozantinib | 3 | ||
| TSC1 | Oncogenic mutations | CNS | Everolimus | 2 |
| RCC | Everolimus | 2 | ||
| TSC2 | Oncogenic mutations | CNS | Everolimus | 2 |
Level 1: FDA-recognized biomarker predictive of response to an FDA-approved drug in this indication
Level 2: Standard care biomarker predictive of response to an FDA-approved drug in this indication or another indication; including those recommended by NCCN, but not FDA recognized as standard of care
Level 3: Evidence of clinical activity in this indication, or another indication
Level 4: Preclinical or biologic evidence of activity
The receptor tyrosine kinases (RTKs) comprise a family of transmembrane (TM) cell surface receptors that transduce extracellular signals internally to promote growth and survival and/or to regulate other cellular phenotypes. Members of this protein family share a similar modular domain structure. Growth factors bind to the extracellular ligand-binding domain of RTKs and induce dimerization of two receptor monomers, juxtaposing the intracellular tyrosine kinase domains of each monomer. This results in transphosphorylation of tyrosine residues within the cytoplasmic domains of the RTK dimer. Following transphosphorylation, a variety of intracellular proteins are recruited to the activated RTK through Src homology 2 (SH2) domains that recognize the phosphotyrosine plus a specific amino acid sequence motif C-terminal to the tyrosine residues.
Over 117 SH2 domains have been characterized, each with unique phosphotyrosine sequence specificities. Each domain is part of a larger adaptor protein involved in transducing extracellular signals to activate, or in some cases suppress, specific intracellular signaling cascades. Thus the complement of signaling pathways that a given RTK regulates is dictated by the profile of phosphorylated tyrosine residues plus flanking amino acids within their intracellular domains. However, more than one adaptor protein can often recognize individual context-dependent phosphotyrosine motifs within an RTK, underscoring how this system is designed to provide both specificity and diversity of intracellular signaling.
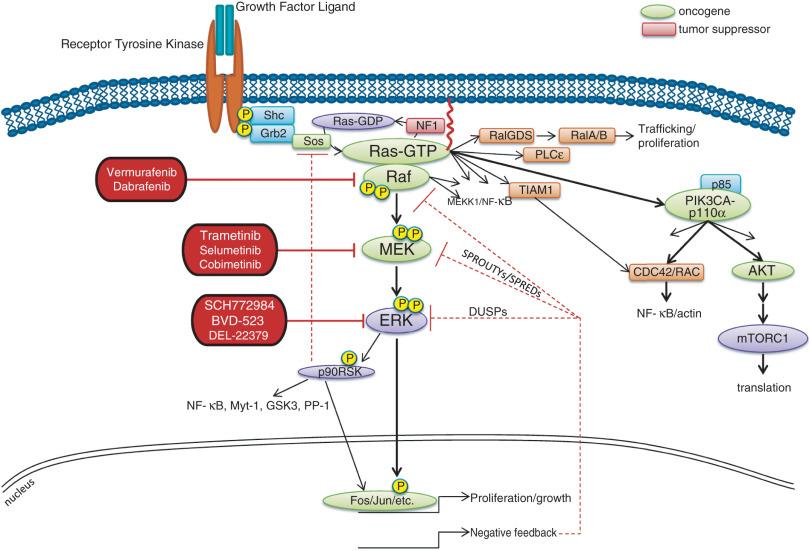
Recruitment of signaling intermediaries to the plasma membrane facilitates their interaction with membrane-bound proteins responsible for stimulating a diverse array of downstream pathways ( Fig. 2.1 ). As an example, the lipid kinase phosphatidylinositol 3-kinase (PI3 kinase), described in more detail in a later section, recognizes and binds to a pattern of phosphorylated tyrosine residues present within multiple activated RTKs through the SH2 domain located in its p85 regulatory subunit. Binding of the p85 regulatory subunit in turn results in activation of its kinase activity. Approximately 20 classes of RTKs have been defined based on growth factor specificity. This section will focus on those RTK classes for which specific cancer therapies exist or are in development.
Historically, the growth factors that stimulate RTKs were first discovered, followed by the structural and functional characterization of the RTKs themselves. Epidermal growth factor (EGF) was initially purified from mouse submaxillary glands in 1962 by Stanley Cohen and was found to stimulate premature eyelid opening and incisor eruption, phenotypes that suggested a role for EGF in the regulation of cellular proliferation. In 1978, the epidermal growth factor receptor (EGFR) was identified as the cell surface binding site for EGF. Over the next several years, tyrosine phosphorylation was identified in cells, followed by the discovery that the viral Src oncogene, which induces transformation of cells in vitro, is itself a tyrosine kinase, underscoring the potential importance of tyrosine phosphorylation for oncogenesis. Once the complete sequence of the EGFR protein was elucidated in the 1980s, the amino acid sequence of the receptor cytoplasmic domain was found to be similar to Src, suggesting that EGFR also possessed tyrosine phosphorylation activity. The connection between RTK activation and oncogenesis was further solidified when the amino acid sequence of EGFR was found to be homologous to the avian erythroblastosis virus erbB oncogene, which, when infected into chicken red blood cell precursors, is sufficient to induce erythroleukemia. The erbB oncogene encodes a TM protein that lacks the extracellular ligand binding domain of EGFR but possesses a cytoplasmic kinase domain that, when expressed in cells, can signal in a growth factor–independent manner. Subsequent studies have since identified within human cancers numerous alterations of EGFR and other RTKs that enhance proliferation without the need for growth factor stimulation.
The EGFR class of RTKs comprises four receptor proteins encoded by four genes (in parentheses): EGFR (ERBB1), HER2/Neu (ERBB2), HER3 (ERBB3), and HER4 (ERBB4). EGFR binds to and is activated by a number of ligands, including EGF, transforming growth factor–α (TGF-α), HB-EGF, amphiregulin, betacellulin, epiregulin, and epigen. Growth factor binding promotes either homodimerization or heterodimerization with other HER family members, followed by transphosphorylation. A ligand for HER2 has not yet been identified; instead, HER2 is activated through heterodimer formation with one of the other three ligand-bound receptors. Notably, HER2 is the preferred dimerization partner for EGFR, and EGFR-HER2 heterodimers are more stable than EGFR homodimers, remaining at the cell surface for a longer duration and undergoing endocytosis at a lower rate than EGFR homodimers. Furthermore, HER2 reduces the dissociation rate of EGF from EGFR, allowing for a more sustained period of EGF-induced signaling. The EGFR and HER2 components of the EGFR-HER2 heterodimer are also more likely to be recycled back to the cell surface than EGFR homodimers, which are more readily targeted for degradation. In addition, HER2-HER3 heterodimers possess the most potent mitogenic activity among the heterodimer and homodimer HER kinase combinations. In contrast to the other HER kinase family members, HER3 does not have intrinsic kinase activity and preferentially forms heterodimers with HER2. The ligands for HER3 and HER4 are the neuregulins, including heregulin.
A number of tumor types frequently exhibit alterations within the EGFR family of RTKs. Sustained activation of these pathways can result in oncogene- or pathway-addicted tumors, and selective HER kinase inhibitors are now a component of the standard treatment of several malignancies. Alterations that affect RTK activity include mutations that result in constitutive activation of the tyrosine kinase; overexpression of the receptor, often due to gene amplification; and elevated levels of RTK ligands that stimulate signaling. EGFR mutations are found in 10% to 25% of non–small cell lung cancers (NSCLCs), with variation in the frequency of such alterations influenced by ethnicity and geographic location; in-frame microdeletions in exon 19 and point mutations in exon 21 (most commonly L858R or L861Q) or exon 18 (G719X) comprise over 80% of these alterations. In glioblastoma multiform (GBM), EGFR mutations, indels (including the EGFRvIII variant in which exons 2 to 7 of the extracellular domain are deleted, generating a ligand-independent, activated protein), amplification, splice variants, and rearrangements occur in 57% of tumors. However, because of the heterogeneity of GBM tumors, targeting EGFR is complicated; EGFR alteration is often concurrent with amplification or mutation of another RTK such as PDGFR, MET, or FGFR, or the presence of EGFRvIII on extrachromosomal DNA, or activation of IDH1. Overexpression of wild-type EGFR as a result of gene amplification has been observed in NSCLC and breast, gastric, colorectal, and head and neck cancers, and less commonly in other tumor types.
Up to 30% of breast cancers display overexpression of HER2, which is an unfavorable prognostic factor, and therapy for these ERBB2 -amplified breast cancers is now distinct from that of breast cancers with normal HER2 expression levels. ERBB2 amplification is also a driving event in gastric and to a lesser extent in bladder, endometrial, and cervical cancer. More recently, activating mutations and in-frame insertions/indels in ERBB2 were found to occur in 1% to 2% of all cancer patients, most commonly in patients with bladder cancer. Mutations in ERBB2 localize to either the extracellular domain, where they are presumed to promote dimer formation, or the kinase domain. In addition, activating mutations in ERBB3 have also been identified in bladder, colon, and gastric cancers.
Numerous targeted agents have been developed that selectively inhibit EGFR-induced signaling (see Table 2.1 and Fig. 2.2 ). Cetuximab, a chimeric monoclonal antibody that binds to the extracellular domain of EGFR and competitively inhibits ligand binding, thereby preventing receptor activation, is approved for the treatment of KRAS wild-type colorectal and head and neck cancers. Panitumumab, another human monoclonal anti-EGFR antibody, is also approved for KRAS wild-type metastatic colorectal cancer.
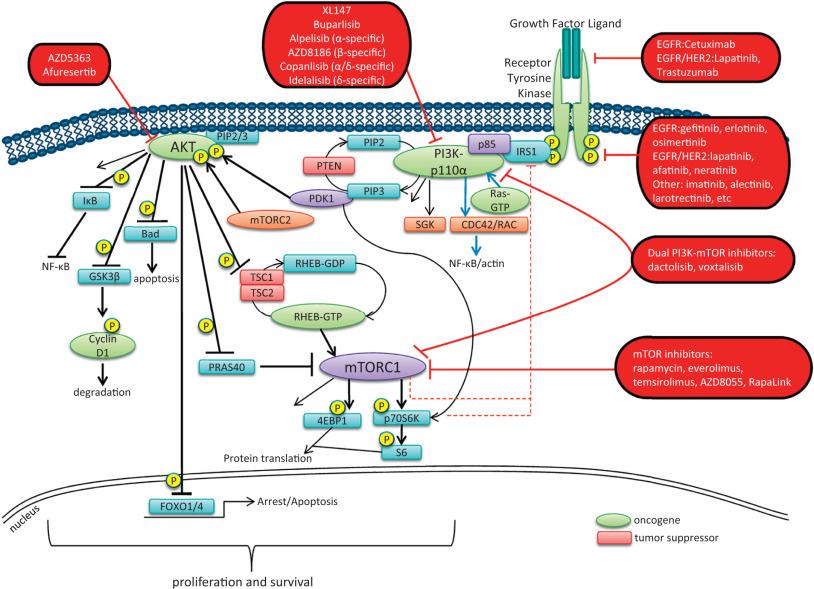
The first-generation reversible EGFR tyrosine kinase inhibitors gefitinib and erlotinib, as well as the second-generation irreversible inhibitor afatinib, are FDA approved for the treatment of NSCLC, with greatest efficacy in patients with EGFR mutations or in-frame deletions. A second site mutation in EGFR (T790M) is a common mechanism of acquired resistance to first generation EGFR inhibitors. Osimertinib (AZD9291), a third-generation EGFR inhibitor, is highly active in patients with NSCLC in which resistance is mediated by the EGFR T790M mutation and is now FDA approved for this indication. The development of osimertinib and the fourth-generation EGFR inhibitor EAI045 highlights how studies of acquired resistance can lead to the rational development of more effective kinase inhibitors.
ERBB2 amplification strongly correlates with HER2 protein overexpression, and the presence of either marker predicts for trastuzumab response in certain cancers. Trastuzumab, a humanized antibody that binds to the extracellular domain of HER2, has been FDA approved for the treatment of breast and esophagogastric cancers displaying HER2 overexpression. In patients with breast and gastric cancers, trastuzumab has modest activity when administered as single-agent therapy and is most commonly used in combination with chemotherapy. The combination of docetaxel, trastuzumab, and pertuzumab, an antibody that binds to a different HER2 epitope (the dimerization domain) than trastuzumab and results in impaired dimer formation, is also approved for breast cancer.
Although the introduction of trastuzumab has resulted in a significant improvement in the survival of patients with HER2-overexpressing breast cancers, drug resistance remains a major clinical problem. Potential resistance mechanisms include concomitant overexpression of other HER kinase family members and/or ligands, PTEN loss, and the expression of a truncated HER2 protein lacking the extracellular antibody binding site. Additional HER2-directed agents include the tyrosine kinase inhibitors lapatinib and neratinib (see Table 2.1 ). Lapatinib is FDA approved for use in combination with capecitabine in patients with HER2-overexpressing advanced or metastatic breast cancer that has progressed on prior therapy with trastuzumab and certain classes of chemotherapy. The combination of lapatinib and trastuzumab is also FDA approved in HER2 -amplified breast cancer. Lapatinib also received accelerated approval for use in combination with the aromatase inhibitor letrozole. Clinical efficacy has been reported with lapatinib in HER2 -mutant NSCLC. The irreversible pan-HER kinase inhibitor neratinib has shown promising clinical activity in patients with ERBB2 -amplified and ERBB2 -mutant breast tumors, but also other cancer types.
The insulin and insulin-like growth factor 1 (IGF1) receptor family is dysregulated in multiple malignancies. The insulin receptor exists as two isoforms encoded by splice variants of the same gene. Each isoform can dimerize with the other (forming hybrid dimers) or with itself. The IGF1 receptor (IGF1R) can dimerize with either of the insulin receptor isoforms or with itself, resulting in six different dimer combinations. The insulin receptor is stimulated by insulin or insulin-like growth factor 2 (IGF2), whereas IGF1R can be activated by either IGF1 or IGF2. Both of these latter ligands can stimulate IGF1R in an autocrine fashion or can be elaborated from distant sites. Circulating IGF binding proteins have a similar affinity for IGF1 and 2 as IGF1R does and therefore compete for binding to both ligands, thus titrating the amount of free ligand available for IGF1R stimulation. IGF binding protein proteases provide an additional mechanism for controlling ligand levels by increasing the half-life of free ligand available for receptor binding.
After ligand binding, IGF1R dimerizes and undergoes transphosphorylation, leading to activation of downstream signaling pathways, including both the Ras-Raf-MAPK and the PI3 kinase-AKT-mTOR cascades (see individual sections later in the chapter; see also Fig. 2.1 ). Specifically, insulin receptor substrate 1 (IRS1) binds to a phosphotyrosine motif on IGF1R via its SH2 domains and is phosphorylated by IGF1R. It subsequently recruits PI3 kinase to the plasma membrane, which converts phosphatidylinositol-4,5-bisphosphate (PIP 2 ) to phosphatidylinositol-3,4,5-triphosphate (PIP 3 ), subsequently resulting in AKT and mTOR pathway activation.
Activating mutations of IGF1R do not appear to be common in human cancer. However, amplification of the IGF1R gene locus has been identified in some colon, pancreas, and lung cancers. Sarcomas often exhibit either increased expression of the IGF1 and IGF2 ligands or decreased IGFBP-3 expression (Ewing sarcoma), which results in increased IGF1 levels in the tumor microenvironment. Gastrointestinal stromal tumors (GISTs) lacking c-KIT and platelet-derived growth factor receptor (PDGFR) mutations also commonly harbor IGF1R amplification. AMG479, a monoclonal human antibody targeting IGF1R, has shown promising antitumor activity in patients with Ewing sarcoma.
Activating kinase domain point mutations and gene rearrangements of the insulin receptor family members anaplastic lymphoma kinase (ALK) and ROS1 play driving roles in many cancers, most notably lymphomas, neuroblastoma, NSCLC, and thyroid cancer. Chromosomal translocations involving ALK and at least 22 5′ fusion partners have been identified, which dictate spatial and temporal expression of the ALK fusions, and likely their function and tumorigenic potential. In NSCLC, the EML4 gene is the preferred translocation partner, resulting in the expression of an EML4-ALK fusion protein in 4% to 6% of patients. Notably, EML4-ALK fusions are found in a mutually exclusive pattern with EGFR kinase domain mutations, suggesting that they have overlapping downstream effects. ROS1 gene rearrangements are also found in a minority of NSCLC patients with binding partners including SLC34A2 and CD74. Crizotinib, an inhibitor of the ALK, ROS1, and MET tyrosine kinases (see Table 2.1 ), is now FDA approved for use in NSCLC patients with ALK or ROS1 fusions (see Table 2.1 ), although acquired resistance mutations in ALK (of note, C1156Y and the gatekeeper mutation L1196M) commonly develop. Newer ALK inhibitors including alectinib and ceritinib that are either more potent or more selective for ALK have significant clinical activity in patients with acquired resistance to crizotinib and are FDA approved for this indication. In addition, brigatinib, a dual inhibitor of ALK and EGFR, was granted accelerated FDA approval in 2017 for patients with metastatic NSCLC and in patients with ALK alterations who progressed on crizotinib.
Platelet-derived growth factor (PDGF) is the ligand for PDGFRs, which stimulate the proliferation and migration of mesenchymal cells, such as oligodendrocyte precursors, vascular smooth muscle cells, and pericytes during embryonic development. PDGF signaling is also implicated in organ development, including lung and intestinal epithelial folding and glomerular capillary tuft formation. Furthermore, PDGFs promote angiogenesis, wound healing, and erythropoiesis. Aberrations in the PDGFR pathway result in uncontrolled proliferation and enhanced angiogenesis.
Four isoforms of PDGF have been identified: PDGFA, PDGFB, PDGFC, and PDGFD. These isoforms are activated by proteolytic cleavage and assemble into five homodimeric or heterodimeric combinations that bind to and stimulate either PDGFRα or PDGFRβ. PDGFRα homodimers inhibit chemotaxis, whereas PDGFRβ homodimers and α/β heterodimers stimulate chemotaxis within fibroblasts and smooth muscle cells. Angiogenic endothelial cells recruit PDGFRβ-positive pericytes to cover blood channels and aid in their maturation and stabilization through secretion of PDGFβ. Following dimerization and transphosphorylation, PDGFRs activate signal transduction pathways through recruitment of adaptor proteins containing SH2 domains, most notably the Grb2 protein, which in turn binds the guanine nucleotide exchange factor (GEF) Sos, which subsequently activates Ras. In addition, phosphorylated tyrosine residues serve as docking sites for SH2 domain–containing kinases, including PI3 kinase, phospholipase-Cγ, and Src, as well as the tyrosine phosphatase SHP2 and the STAT transcription factor family.
Alterations in PDGFR signaling in cancer include excess autocrine secretion of PDGF (glioblastoma, sarcomas), gain-of-function mutations that cause constitutive tyrosine kinase activation (GISTs), translocation of either the PDGF or PDGFR gene (dermatofibrosarcoma protruberans, chronic myelomonocytic leukemia, hypereosinophilic syndrome), and PDGFR gene amplification (glioblastoma). PDGFRα mutations are found in approximately 10% of KIT wild-type GISTs and are sensitive to imatinib, a tyrosine kinase inhibitor of KIT, BCR-ABL, and PDGFRs, which is standard of care in this setting (see Table 2.1 ). The D842V mutation comprises approximately two-thirds of PDGFRα activating mutations, and confers resistance to imatinib. Notably, the second-generation inhibitor dasatinib is effective in preclinical models of imatinib-resistant GIST. Dermatofibrosarcoma protuberans is a rare, low-grade cutaneous sarcoma that harbors a chromosome 17;22 translocation that fuses portions of the COL1A1 (collagen 1A1) gene and PDGFB , resulting in overexpression of PDGF-β and subsequent stimulation of PDGFR signaling. Twenty other fusions partners have been identified in PDGFβ rearrangements, including ETV6 and EBF1. Imatinib has shown significant benefit in patients with recurrent or metastatic dermatofibrosarcoma protuberans, myelodysplasia, and myeloproliferative neoplasms and is FDA approved for these indications.
The KIT gene is a member of the type III RTK family, which includes PDGFR and FLT3 (see later). It was first identified as the human homologue of the viral oncogene v-Kit responsible for the Hardy-Zuckerman IV feline sarcoma virus. Mutation of KIT or its ligand, stem cell factor (SCF), in mice induces coat color abnormalities (“white spotting”), anemia, and mast cell deficiencies, suggesting that it plays a role in hematopoiesis and melanogenesis. Furthermore, the KIT protein was discovered as a cell surface receptor in acute myeloid leukemia (AML). KIT expression is mainly restricted to mast cells, hematopoietic cells, germ cells, melanocytes, and the interstitial cells of Cajal (ICCs) in the gut. SCF and KIT integrate signals that lead to mitogen-activated protein kinase (MAPK), PI3K, and SRC pathway activation, and mediate critical survival and proliferation cues to distinct hematopoietic lineages, including the bone marrow and progenitor cells.
Hot-spot mutations in exons 9 and 11 of KIT have been identified in several tumor types, including GIST, melanomas, and germ cell tumors. In GIST, 85% of tumors have activating KIT mutations that drive the transformation of precursors of ICCs. Imatinib and sunitinib inhibit KIT and PDGFR, among other kinases, and are FDA approved for use in patients with KIT mutant GIST (see Table 2.1 ). Regorafenib is approved for patients with imatinib- or sunitinib-refractory GIST. Imatinib has also been shown to induce tumor regression in patients with KIT -mutant or KIT -amplified melanoma. Second site mutations in KIT, typically in exon 17, are a mechanism of acquired resistance to imatinib therapy in patients with GIST. Novel agents that retain activity in the setting of an exon 17 KIT mutation are now in clinical testing (clinical trial NCT02401815). New areas of investigation into mutant KIT therapies in GIST include blocking mutant KIT subcellular localization to the Golgi and combination of FGFR3 and KIT inhibitors to quench pathway cross talk.
The FMS-like tyrosine kinase 3 receptor (FLT3), a third member of the RTK class that includes PDGFR and KIT, is involved in the development of normal hematopoietic cells. It contains an extracellular region composed of five immunoglobulin (Ig) domains, TM and juxtamembrane domains, and two cytoplasmic tyrosine kinase domains that transmit proliferative signals through the RAS/MAPK, PI3K/AKT, and STAT5 pathways. Two main FLT3 alterations are common in hematopoietic malignancies, namely in approximately 30% of AMLs. First, internal tandem duplication (ITD) within exons 14 and 15 of the FLT3 gene (FLT-ITD) interferes with the negative regulatory function of the juxtamembrane segment. This duplication results in ligand-independent activation of FLT3 and is associated with a poor prognosis in patients with AML. Second, kinase domain mutations at or near D835 in the activation loop of FLT3 disrupt autoinhibitory interactions and render the kinase open and active. The clinical activity of FLT3 inhibitors has been modest to date, although responses appear to be more common in patients with FLT3/ITD AML. TAK-659, a reversible dual Syk/Flt inhibitor, showed early clinical activity in numerous lymphoma subtypes and AML. Sorafenib has been shown in preclinical in vitro studies, mouse models, and in a phase I study of AML patients to reduce leukemia burden and block signaling selectively in FLT3-ITD versus FLT-wt settings. Interesting to note, resistance to FLT3 inhibition in such patients is associated with selection for secondary mutations within the tyrosine kinase domain of FLT3, suggesting a central role of FLT3 in AML pathogenesis.
Fibroblast growth factor receptors (highly conserved FGFR1, FGFR2, FGFR3, and FGFR4; and FGFRL1/FGFR5, which lacks a kinase domain) comprise a family of RTKs that regulate cell proliferation, differentiation, and migration as well as selective apoptosis during embryogenesis. The FGFRs are composed of an extracellular ligand-binding domain, a hydrophobic TM region, and an intracellular tyrosine kinase domain. The extracellular domain is organized into three Ig domains; differential splicing of the second half of the third Ig domain dictates tissue-specific expression of the receptor.
Fibroblast growth factors (FGFs) are protein ligands that bind to the extracellular domain of the FGFRs in combination with specific heparan sulfate glycosaminoglycans inducing FGFR dimerization and transphosphorylation of intracellular tyrosine residues. Eighteen FGFs have been identified, and specificity for FGFRs is based on numerous factors, including tissue-specific FGF ligand and receptor expression, the presence of cell surface molecules that facilitate the interaction between individual FGF ligands and receptors, and the differential binding capability of the ligands themselves for specific FGFRs. Subsequent stimulation of the tyrosine kinase domain leads to phosphorylation and activation of multiple downstream signaling proteins in the same manner as described earlier for other RTKs. Unique to the FGFR signaling complex is FGFR substrate 2 (FRS2), an adaptor protein that binds to specific phosphotyrosines on the intracellular domain of active FGFR dimers. FRS2 is itself phosphorylated by FGFRs and serves as a docking site for the Grb2-Sos adaptor complex, which activates the Ras/Raf/MAPK pathway. Phosphorylated FRS2 also recruits Grb2-associated binding protein 1 (GAB1), which activates PI3 kinase. In addition, phospholipase-Cγ binds to phosphorylated FGFR dimers via an SH2 domain, leading to its activation and the cleavage of phosphatidylinositol 4,5-bisphosphate (PIP 2 ) to form inositol 1,4,5-triphosphate (IP 3 ) and diacylglycerol (DAG).
Germline mutations of the FGFR genes are the basis of a spectrum of skeletal developmental disorders that are thought to derive from premature differentiation and growth restriction of chondrocytes resulting from dysregulated FGFR pathway activation. FGFR signaling is dysregulated in cancer by multiple mechanisms including mutational or translocation-induced activation of FGFRs, gene amplification of receptors, and abnormal ligand regulation. For example, autocrine and paracrine FGF ligand secretion with resultant pathway activation has been reported to occur in a subset of melanomas and prostate cancers, respectively. FGFR1 amplification occurs in approximately 17% of squamous cell lung cancers and 6% of small cell lung cancers (SCLCs). Approximately 10% of diffuse-type, aggressive gastric cancers display FGFR2 gene amplification, and cell lines with this amplification show ligand-independent pathway activation and sensitivity to selective FGFR inhibitors. Whereas FGFR1 mutations are rather rare, FGFR2 mutations are found in approximately 10% of endometrial cancers. FGFR3 mutations occur in up to 75% of non–muscle invasive bladder cancers and 15% of patients with advanced urothelial tumors. Activating mutations within FGFR3 result in constitutive receptor dimerization and subsequent signaling. Unlike EGFR-activating mutations, which predominantly affect the tyrosine kinase domain of the receptor, FGFR3 mutations are commonly located within the extracellular domain (R248, S249) and TM segment (G370, Y373) and promote ligand-independent receptor dimerization through formation of an aberrant disulfide bridge between two receptor monomers. Chromosomal rearrangement of FGFRs have been identified using next-generation sequencing. Up to 15% of multiple myelomas harbor an intergenic 4;14 translocation between the FGFR3 gene and the Ig heavy chain locus, which places FGFR3 expression under the highly active heavy chain promoter. More recently, translocations involving FGFR2 have been reported in cholangiocarcinoma and more rarely in other cancers, whereas FGFR3 fusions are most common in glioblastoma and bladder cancers but also are found rarely in other solid tumor types. The FGFR3-TACC3 constitutively active fusion protein has been characterized to induce aneuploidy by disrupting proper chromosomal segregation.
Multiple FGFR inhibitors are currently being tested in early-phase clinical trials, but the majority of these compounds are multitargeted tyrosine kinase inhibitors, many of which also potently inhibit members of the VEGFR and PDGFR families. The close structural similarity between these RTKs has made development of FGFR-selective inhibitors challenging, although several such drugs are now in early clinical testing, such as BGJ398, AZD4547, JNJ-42756493, and Debio1347 (see Table 2.1 ). On-target hyperphosphatemia resulting from FGFR1 inhibition is a primary toxicity with this class of agents, suggesting that the development of isoform-selective FGFR inhibitors may be a more rational approach for patients whose tumors are driven by mutations or translocations in FGFR2 and FGFR3. FGFRs are located on the cell surface and thus may also be susceptible to monoclonal antibody mediated inhibition similar to trastuzumab-mediated inhibition of HER2. FGFR ligand traps are also in development.
The RET (Rearranged During Transfection) gene encodes three alternatively spliced isoforms (RET9, RET43, and RET51) which are TM RTKs that contain four cadherin-like extracellular repeats important for dimerization, a cysteine-rich juxtamembrane region critical for ligand binding and conformation, and an intracellular kinase domain. RET9 and RET51 are highly conserved in all vertebrates and play important roles in the normal development and maintenance of many tissues, including the kidney, spermatogonial stem cells, and the enteric nervous system. RET is expressed predominantly on the surface of neural crest tissues, and glial-derived neurotrophic factors (GDNFs) such as neurturin, artemin, and persephin serve as ligands for RET. GDNFs initially bind to their cognate coreceptors, the GDNF receptors (GFPα1 to GFPα4), on the cell surface, which recruits RET into lipid raft membrane domains and causes conformational changes via the cadherin-like moieties, dimer formation, and then subsequent transphosphorylation of tyrosine residues and kinase activation. Y1062 is the common docking site for all three RET isoforms and serves to recruit many adapters including SHC1, FRS2, IRS1/2, DOK, and JNK. Phosphorylation of Y752 and Y928 binds STAT3, whereas other phosphorylated residues are recognized by Src, resulting in activation of focal adhesion kinase (FAK), which promotes cell migration and metastatic spread. In addition, the MAP kinase, PI3 kinase/AKT, and phospholipase-Cγ pathways can be activated by RET to promote cellular proliferation and survival.
Germline loss-of-function RET mutations occur in Hirschsprung and CAKUT (congenital anomalies of the kidney and urinary tract) disease, which causes abnormalities of the developing gut and kidneys, respectively. Conversely, germline activating RET mutations are the basis for the multiple endocrine neoplasia type 2 (MEN2) syndromes. Patients with MEN2 develop familial medullary thyroid carcinomas and other cancers. MEN2A is mainly driven by mutations in six cysteine resides in the RET extracellular domain (C609, C611, C618, C620, C630, and C634), whereas the kinase domain mutations M918T or A883F are associated with MEN2B. Sporadic medullary thyroid carcinomas are much more common, and up to 60% of such tumors harbor somatic mutations in RET , notably G691S, which are thought to be a driver alteration in this disease. Furthermore, RET gene rearrangements with numerous fusion partners, including CCDC6 and NCOA4, termed RET-PTC1 and RET-PTC3, respectively, occur in 20% to 40% of papillary thyroid carcinomas (PTCs) and often occur as a consequence of high doses of radiation.
RET inhibitors have shown significant antitumor activity in patients with medullary thyroid cancer. Vandetanib, an oral inhibitor of RET, EGFR, and VEGFR, is FDA approved for the treatment of patients with advanced medullary thyroid cancer. A randomized, placebo-controlled phase III study of cabozantinib, an oral, multitargeted TKI that inhibits RET, VEGFR2, and MET, was also recently conducted in patients with unresectable, locally advanced, or metastatic medullary thyroid carcinoma. This trial documented a statistically significant improvement in median progression-free survival with cabozantinib as compared with placebo (11.2 months versus 4.2 months in placebo arm, P < .0001). More recently, cabozantinib was FDA approved for the treatment of renal cell carcinomas that progressed on antiangiogenic therapy, although the activity of cabozantinib in this context may not be attributable to its inhibition of RET. Several RET inhibitors have, however, shown promising clinical activity in patients with NSCLC treated with RET fusions (see Table 2.1 ).
Six vascular endothelial growth factor (VEGF) ligands have been identified: VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placental growth factors 1 and 2. VEGF-A has four isoforms produced by alternative gene splicing, with the 165–amino acid length isoform playing a central role in tumor angiogenesis. Specifically, VEGF-A enhances vascular permeability and stimulates endothelial cell proliferation, resulting in new blood vessel formation. Vascular endothelial growth factor receptors (VEGFR1 to VEGFR3) are RTKs that possess a modular structure consisting of an extracellular domain with seven Ig-like regions, a TM domain, and an intracellular tyrosine kinase domain. VEGF-A, VEGF-B, and placental growth factor all bind VEGFR1 (also known as FLT1), but the exact role of VEGFR1 in tumor angiogenesis has yet to be fully elucidated. In some settings it may act as a decoy receptor that prevents ligand-mediated stimulation of VEGFR2 (also known as FLK1/KDR). VEGFR2 has been implicated in the development of vasculature during development and is considered the primary receptor through which VEGF exerts its angiogenic effects in endothelial cells. Binding of ligand to VEGFR2 results in receptor dimerization and transphosphorylation followed by activation of multiple mitogenic signal transduction cascades. More recently, VEGF has been implicated in many angiogenesis-independent roles including regulation of immune cells in the tumor microenvironment, fibroblasts in the tumor stroma, and cancer stem cells. VEGF can also bind and signal through a class of TM glycoprotein coreceptors called neuropilins (NRP1 and NRP2), which are found on tumor cells and can signal along many oncogenic axes including Hedgehog and JNK.
The complex network of cross talk among VEGFs, VEGFRs, and canonic oncogenic pathways makes VEGF and VEGFR critical but elusive targets in cancer therapy. Targeted therapies that inhibit VEGF signaling include antibodies that bind circulating ligand and RTK inhibitors. The humanized monoclonal antibody bevacizumab binds to free VEGF, thereby preventing its association with VEGFRs. This antibody has been FDA approved for use in combination with chemotherapy for patients with several cancers, including metastatic colorectal and nonsquamous NSCLCs. Bevacizumab also has activity in patients with glioblastoma and metastatic renal cell carcinoma, where it is often used in combination with interferon-α (IFN-α). In addition, ramucirumab is a VEGFR2-directed antibody that has received FDA approval with or without chemotherapy in several cancers.
Sorafenib, sunitinib, pazopanib, and axitinib are multitargeted tyrosine kinase inhibitors with nanomolar potency for VEGFR2. Sunitinib is used in the treatment of patients with metastatic renal cell carcinoma, GISTs, and pancreatic neuroendocrine tumors. Sorafenib has been approved for the treatment of liver and renal cell cancers. Although these agents inhibit multiple kinases, their antitumor effects have been attributed primarily to their antiangiogenic activity. More recently, the tyrosine kinase inhibitor pazopanib was approved for the initial treatment of metastatic renal cell carcinoma and in cytokine-pretreated patients, and axitinib was approved in the second-line setting following failure of prior systemic therapy. Lenvatinib, a multitargeted RTK inhibitor that inhibits VEGFR1, VEGFR2, and VEGFR3, has received recent FDA approval for both thyroid cancer (as monotherapy) and renal cell carcinoma in combination with everolimus.
Despite widespread activity in preclinical models, antiangiogenic therapies have shown disappointing activity in several tumor types. A number of resistance mechanisms have been hypothesized to explain the lack of broader clinical activity, including the activation of redundant signaling pathways that promote angiogenesis; the recruitment by tumors of bone marrow–derived endothelial progenitor cells; increased pericyte density around existing blood vessels, which enhances vascular growth and survival; and the ability of tumor cells to invade surrounding stroma to co-opt additional blood supply. A better understanding of these resistance mechanisms may lead to the development of more effective antiangiogenic therapies in the future.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here