Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Patients with systemic lupus erythematosus (SLE) may experience arthritis, serositis, renal involvement, and a wide range of other clinical manifestations with the potential to affect nearly any organ system. These pathologies are driven by chronic inflammation and the presence of autoantibodies, which arise through complex interactions between genetic predisposition, hormonal influences, and environmental exposures. For example, infections commonly lead to transient, low-level production of autoantibodies by stimulating the activation of cytokine pathways, differentiation and activation of immune cells, and exposure to autoantigens. In most individuals, these changes do not lead to autoimmune disease because immunoregulatory mechanisms restore tolerance to self-antigens and suppress inflammation after the infection is cleared. However, in individuals who are genetically susceptible to SLE, exposure to certain pathogens can shift the immune system toward a dysregulated state that permits the onset and progression of autoimmune disease.
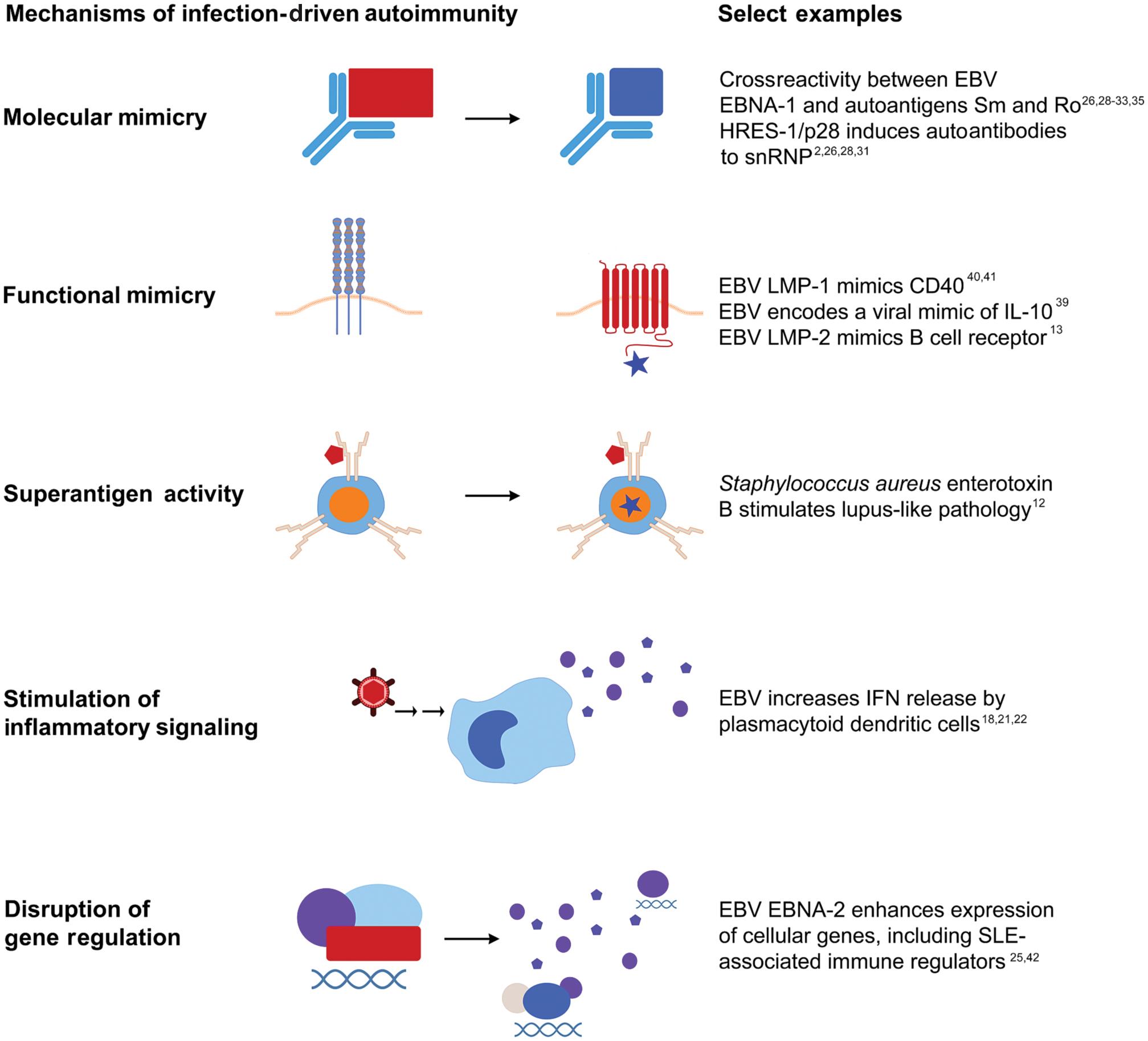
Pathogens promote lupus autoimmunity and clinical SLE through mechanisms such as molecular mimicry, functional mimicry, superantigen activity, and disruption of immunoregulatory mechanisms ( Fig. 24.1 ). Conversely, infections with certain pathogens at certain stages of development may protect against lupus and other autoimmune diseases. Commensal microbes have also been linked with lupus autoimmunity through these mechanisms, particularly molecular mimicry and disruption of immunoregulatory mechanisms. The role of the microbiome in lupus pathogenesis is detailed in Chapter 24.
Several lines of evidence link viral infections with SLE pathogenesis. Case reports have documented at least 25 patients who initially fulfilled the American College of Rheumatology SLE classification criteria after acute viral infection, including 3 after Epstein–Barr virus (EBV) infection, 15 after parvovirus B19 infection, and 6 after cytomegalovirus (CMV) infection. Each of these viruses can also stimulate the production of SLE-associated autoantibodies. Both CMV and parvovirus B19 have been purported to stimulate the onset of SLE because active infections can lead to disease flares in SLE patients and may trigger SLE-like clinical manifestations in previously healthy individuals. Although this chapter focuses on exogenous pathogens, viruses that have integrated into the human genome, such as endogenous retroviruses, are increasingly recognized as fundamental regulators of the innate immune response. Endogenous retroviruses have been linked to select clinical manifestations of SLE, including renal disease and antiphospholipid syndrome in humans, and altered expression of endogenous retroviral elements can regulate the production of autoantibodies and inflammatory mediators.
Finally, pathogenic bacteria may stimulate autoimmunity leading to SLE. Peptides from Vibrio cholerae and Streptococcus agalactiae act as molecular mimics that elicit antibodies reactive to the lupus-associated autoantigen Sm D1. In addition, bacterial superantigens can activate large numbers of T cells, including autoreactive T cells, by avidly binding to class II MHC molecules outside of the conventional antigen-binding clefts. Besides stimulating autoantibody production, the Staphylococcus aureus superantigen and cholera toxin B induce systemic, SLE-like disease in mice. Therefore, infectious agents likely play an important role in SLE pathogenesis.
EBV (human herpesvirus 4) is an attractive candidate in the etiology of SLE because it infects B cells and extensively alters immune regulation. Nearly all individuals worldwide are EBV-infected by early adulthood. After an active EBV infection, EBV remains latent in memory B cells, presenting a lifelong antigenic challenge in most individuals. In addition, infected cells can become immortalized, and EBV has the potential for episodic reactivation.
An association between EBV seropositivity and SLE was first shown in 1971, and many studies have confirmed this association. A recent metaanalysis further confirms this seropositive association, both with VCA IgG [OR 2.06 (95% CI: 1.30–3.26; P = 0.002)] and with EA IgG [OR = 7.70 (95% CI: 4.64–12.76); P < 0.001], as well as an association with DNA-positive EBV infection in SLE patients compared to controls of 3.86 [1.52–9.83] ( P = 0.005). In addition, SLE patients are more likely than controls to show evidence of EBV reactivation, including positivity for viral DNA and antibodies against EBV reactivation proteins. A fundamentally altered response to EBV appears to make SLE patients more susceptible to viral reactivation, leading to active EBV infection and chronic immune stimulation. Compared to healthy individuals, SLE patients exhibit decreased EBV-specific cytotoxic T cell responses, impaired IFN-γ production by EBV-specific CD8 + T cells after EBV stimulation, and markedly increased EBV viral loads.
EBV can act through multiple channels to stimulate lupus autoimmunity and clinical disease in genetically predisposed individuals. For example, EBV stimulates interferon activation and proinflammatory cytokine production, which are considered central to SLE pathogenesis and ongoing pathology ( Chapter 18 on cytokines and Chapter 20 on interferons). In addition, latent EBV infection of B cells can cause transactivation of the human endogenous retrovirus-K18 superantigen, leading to the stimulation of large numbers of cells and differentiation of EBV-infected B cells to memory cells. Further supporting an etiological role for EBV in the development of SLE, some EBV antibodies crossreact with lupus autoantigens, and crossreactive EBV antibodies precede lupus autoimmunity. Finally, EBV has now been linked to SLE and other autoimmune diseases through mechanisms such as molecular mimicry, functional mimicry, and disruption of gene regulatory networks.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here