Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Spinal muscular atrophy (SMA) refers to a group of disorders that affect the lower motor neuron, and a number of genes have been defined that cause SMA. The most common SMA is proximal SMA that maps to chromosome 5q. In contrast, the late-onset distal SMAs (DSMAs), which show considerable overlap with Charcot–Marie–Tooth disease phenotype, are mostly non-5q SMAs. For these disorders many genes have been implicated, including: GARS , DCTN1 , HSPB8 , HSPB1 , BSCL2 , SETX , HSPB3 , DYNC1H1 , REEP1 , and SLC5A7 . Dominant SMAs with proximal weakness have mutations in VAPB , TRPV4 , LMNA , and, most recently, BICD2 . The recessive non-5q SMAs are caused by mutations in IGHMP2 for DSMA1 or SMA with respiratory distress, mutations in Gle1 for severe lethal congenital contracture syndrome 1, which results in loss of motor neurons in the fetus, and lastly the X-linked SMA caused by mutations in Ube1 . Overall, there is no indication of an overlapping function that is disrupted by other causative SMA genes with the exception that, in dominant proximal SMA, the genes VAPB , TRPV4 , and BICD2 may play a role in vesicle release. However, the phenotype of these proximal dominant SMAs is quite different from proximal recessive 5q SMA. For instance, cases with the BICD2 mutation show symptoms at birth with a slow progression, upper motor neuron involvement, and sparing of axial muscle. Therefore it is possible that mutations in different SMA causative genes represent alternate paths to motor neuron destruction. Here we focus on the genetics of 5q SMA.
Proximal 5q SMA is an autosomal recessive disorder and is the most common genetic cause of infant death. Proximal 5q SMA is caused by loss or mutation of the survival motor neuron 1 gene ( SMN1 ) and retention of the copy gene SMN2 . 5q SMA has a frequency of 1/11,000 new births and carrier frequencies range from 1/47 to 1/72, depending on racial group. The SMN1 and SMN2 genes lie in a complex region of the genome that shows a very high divergence among different individuals. Thus, any one particular arrangement of the SMN1 and SMN2 genes does not reflect the arrangement of the genes across the population. Of particular relevance to SMA disease is the inherent copy number variation of SMN1 and SMN2 in the population. The SMN2 copy number varies in SMA individuals and, provided that the SMN2 gene is intact, these extra copies modify the phenotype. An inverse correlation of phenotypic severity to copy number of SMN2 is found in SMA patients. Notably, for many SMAs the mutated gene is expressed in a wide range of tissues and expression is not confined to the nervous system. This is also the case with SMN and as such the reason for selective motor neuron or motor circuit deficit is not known.
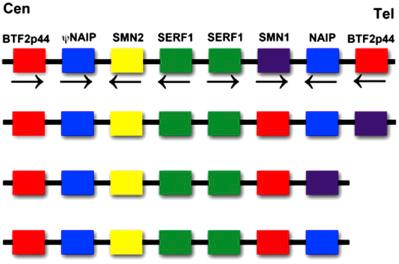
The SMA region is often diagramed as an inverted duplication of SMN1 and SMN2 . This arrangement of genes was identified in a particular yeast artificial chromosome (YAC). However, this region is somewhat more complicated and we now know the arrangement of genes is not the same in all individuals ( Fig. 6.1 ). The gene for SMA was first mapped by linkage analysis to position 5q for all SMA types. Subsequently, as in most positional cloning, closer markers and physical maps of the region between the two flanking markers were then determined. However, the more closely located markers often gave rise to multiple copies in an individual (i.e., greater than two alleles) indicating duplication of the marker. Furthermore, the variation in copy number of these markers between individuals indicated different associated genomic DNA. Some markers indicated reduced gene copy number in many type 1 patients thus suggesting a large deletion in this area. The SMA area was cloned and physical maps were produced utilizing YACs. One difficulty encountered while building these maps was that the YAC clones from this region frequently underwent deletion or some form of rearrangement. This could be easily detected by genotyping of the original cell line and the YAC clones, which most often contained one allele instead of multiple alleles. Thus maps of the SMA region produced from YAC clones with deletions were interpreted as if only one arrangement of multiple chromosomes was possible. Further studies, using pulsed field gel electrophoresis to map the SMA region, revealed marked differences between individuals as different size restriction fragments were identified with probes from the SMA region. Lastly, unequal crossing-over was found in some individuals indicating a noninverted duplication of the SMN genes. Therefore, multiple alternative structures of the SMA region have been identified and as such this area is not always composed of an inverted duplication ( Fig. 6.1 ). One consideration, which even today has not been addressed, is whether particular genomic structures are more prone to new mutations. The rate of new mutation has been reported to be 2%. However, considering the mutation rate by itself, then this should lead to an exceptionally high rate of disease alleles for an autosomal recessive disorder; yet, this is not the case. The most likely reason for this is the presence of an equal back-mutation rate of SMN2 exchange or conversion to SMN1 . Due to the complex nature of this region, it is often incorrectly represented in sequence databases and caution should be applied when extracting data from these sources.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here