Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Specific genes implicated in causing each major form of skin cancer have been identified through genetic studies on hereditary and/or sporadic skin cancer. Their role in promoting cutaneous neoplasia is supported and confirmed by functional studies in animal model systems.
Defects in the CDKN2A tumor suppressor locus are associated with both familial and sporadic cutaneous malignant melanoma and may cooperate with RAS or RAF proto-oncogene activation to promote tumor formation.
Mutations resulting in RAS proto-oncogene activation may cooperate with inactivation of either CDKN2A or p53 tumor suppressor genes in causing cutaneous squamous cell carcinoma.
Defects in the PTCH gene have been implicated in both hereditary and sporadic basal cell carcinoma and mutations in genes encoding other components of the SHH signaling pathway have been associated with sporadic tumors. Defects in the p53 tumor suppressor gene are common in basal cell carcinoma as well.
Although significant advances have been made in identifying genes associated with skin cancers, additional yet-to-be-identified genes likely contribute to the pathogenesis of each major form of skin cancer.
Tumorigenesis is a multi-staged process that derives from a series of genetic alterations, some of which are acquired and others inherited. Genetic aberrations that result in tumor formation alter basic cellular processes, including cell differentiation, cell cycle regulation and cell death. Significant progress has been made over the past two decades in identifying genes associated with specific cancers. The study of hereditary and sporadic skin cancers has led to the identification of numerous genes critical to tumorigenesis. Functional studies using in-vitro and in-vivo models have verified the critical role these genes play in tumor formation.
Cancer-associated genes fall into two general categories: proto-oncogenes and tumor suppressor genes ( Fig. 2.1 ). Proto-oncogenes, such as RAS and RAF , normally regulate cell proliferation or survival. However, upon mutation, proto-oncogenes may be activated to become oncogenes, which allows them to bypass regulatory mechanisms that normally prevent their function in an uncontrolled manner. An activating mutation in just one allele is typically sufficient to contribute to tumorigenesis. By contrast, tumor suppressor genes, such as those encoded in the CDKN2A locus and the TP53 gene, normally inhibit cell cycle progression and proliferation. Inactivation of both alleles of such genes, through mutation, deletion or silencing, is typically required to lose suppressor function and permit tumor formation.
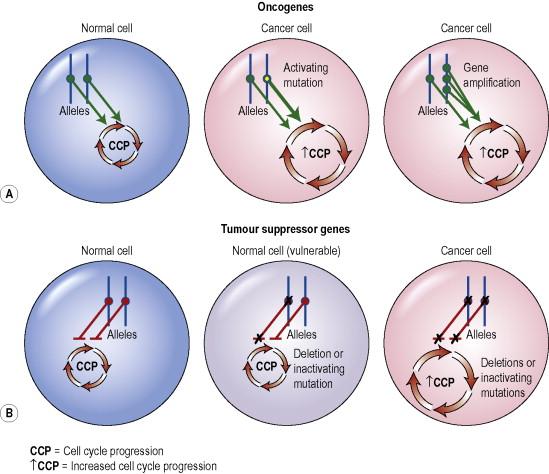
Another form of regulation of both proto-oncogenes and tumor suppressor genes is epigenetic modification that results in the enhancement or silencing of gene expression. These modifications do not change the DNA sequence of the genes but rather affect their expression by covalently modifying either DNA-associated proteins, such as histones, or the DNA itself. Patterns of epigenetic changes linked with specific cancer-associated genes are being established.
In addition, mRNA stability can further be regulated by microRNAs, single-stranded RNA molecules that are 21 to 23 nucleotides in length. MicroRNAs are predicted to regulate the mRNA stability of up to 33% of all genes. They function by encoding complementary sequences to their target genes such that upon binding, the target mRNA is either degraded or its translation to protein is inhibited. MicroRNAs associated with specific cancers are beginning to be identified and functionally characterized. This chapter reviews and summarizes the current understanding of the genetic basis of the three predominant forms of skin cancer: basal cell carcinoma, squamous cell carcinoma, and melanoma.
The most common human malignancies are non-melanoma skin cancers, and among these cancers, basal cell carcinoma (BCC) is the most common type. Insights into the molecular pathogenesis of basal cell carcinomas were originally derived from the genetic analysis of kindreds with basal cell nevus syndrome (BCNS; Gorlin syndrome, OMIM 109400). This syndrome is inherited in an autosomal dominant manner and is characterized by the development of hundreds of BCCs, which can be generalized but are often concentrated in sun-exposed areas. Further, individuals with BCNS demonstrate an increased sensitivity to ionizing radiation, with BCCs developing within radiation ports. Other features of BCNS include palmoplantar pits, odontogenic cysts, calcification of the falx cerebri, skeletal abnormalities, and the development of medulloblastoma.
Linkage analysis of BCNS identified a chromosomal locus at 9q22.3, a locus also found to be deleted in sporadic BCCs. Subsequently, mutations in PTCH1 , the human homolog of the Drosophila patched gene located at 9q22.3, were identified. PTCH1 encodes a transmembrane receptor for the hedgehog family of soluble effector proteins.
The hedgehog signaling pathway is critical to tissue development and homeostasis. First identified in the fruit fly Drosophila melanogaster , hedgehog was found to be one of a set of genes critical to establishing anterior and posterior polarity within the developing fly; flies without hedgehog were found to be shorter than wild-type flies. In vertebrates, hedgehog plays a role in neural tube development. Hedgehog is a secreted lipoprotein that has three mammalian orthologs: sonic hedgehog, Indian hedgehog, and desert hedgehog. Sonic hedgehog signals through the patched family of receptors.
The PTCH1 gene encodes the 12-span transmembrane protein patched that is the receptor for sonic hedgehog. When sonic hedgehog binds patched, the constitutive inhibition of the G-protein receptor smoothened is released, resulting in the release of the Gli (glioma-associated oncogene) transcription factors (Gli1, Gli2, Gli3) from a cytosolic inhibitory complex. This leads to nuclear localization of Gli and a subsequent signaling cascade, which includes a feed-forward induction of PTCH1 ( Fig. 2.2 ). Gli proteins are members of the Kruppel family of zinc finger transcription factors and have been found to mediate activation (Gli1, Gli2) and inhibition (Gli3) of transcription of numerous genes. Tumor-promoting Gli targets include PDGFRα , WNT and IGF2 . Recent studies using small alkaloid molecules that bind smoothened and inhibit the downstream activation of the hedgehog pathway have demonstrated significant antitumor effects against BCCs and may be useful therapeutic agents for treating patients with BCNS or locally advanced BCCs.
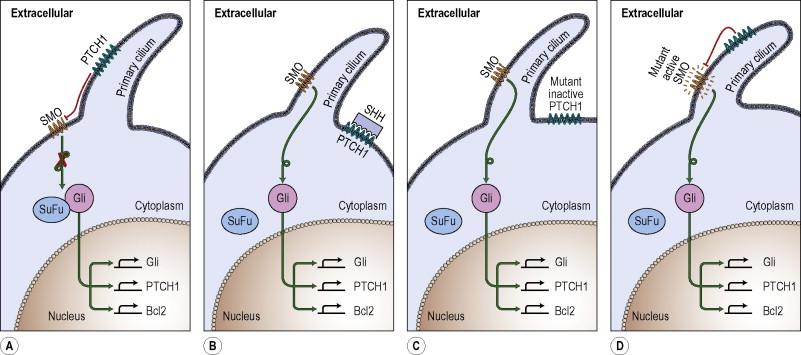
The protein that holds Gli in an inhibitory complex is SUFU, a human homolog of the Drosophila suppressor of fused. As would be predicted, loss of SUFU results in a phenotype consistent with constitutive Gli activation. Mice that are Sufu +/− develop odontogenic cysts and basaloid epidermal proliferations. In human studies, mutations in SUFU have been found in children with medulloblastomas; SUFU mutations have not yet been identified in BCCs.
The hedgehog pathway has been found to rely on the primary cilium during development. Primary cilia are cellular organelles that are present on most cells of the body and play a critical role in intercellular and environmental communication. When hedgehog is present, PTCH1 relocates from the primary cilium to endosomes; conversely, smoothened relocates from intracellular vesicles to the primary cilium. Intriguingly, recent studies in murine models of tumorigenesis have demonstrated that when the primary cilium is absent in cells, activated SMO fails to induce expression of hedgehog target genes and to generate tumors.
PTCH2 encodes a homolog of PTCH1 with ~73% amino acid similarity to PTCH1 . The role of PTCH2 remains undefined; a murine knockout of Ptch2 results in mice with no increased susceptibility to developing tumors. However, crossing Ptch1+/− with Ptch2−/− mice resulted in a higher incidence of tumors and a broader spectrum of types of tumors than in Ptch1+/− mice, suggesting that patched-2 may complement the tumor suppressor role of patched-1. In support of this hypothesis, Ptch2 expression levels have been found to be increased in medulloblastomas in which Ptch1 expression is reduced. In humans, mutant PTCH2 was found to be associated with nevoid basal cell carcinoma syndrome in a Han Chinese kindred.
Like BCCs, medulloblastomas have been found to develop as a result of mutations within the hedgehog signaling pathway. Recently, a microRNA family, the miR-17~19 cluster family, was found to be overexpressed in human medulloblastomas, and forced expression of these microRNAs in Ink4c−/− ; Ptch1+/− mice resulted in the development of medulloblastomas. Whether these microRNAs or others will play a role in the pathogenesis of BCCs remains unknown.
The pathways through which hedgehog signaling induces tumorigenesis remain unclear. Forced expression of SHH was demonstrated to downregulate the expression of p21CIP1, a cell cycle inhibitor. In a murine model, elimination of Ptch1 from mouse skin resulted in basal cell-like tumors that were found to have accumulated the cell cycle regulators cyclin D1 and B1 within their nuclei. Further, patched-1 has been demonstrated to bind directly to cyclin B1 and thus prevent its translocation in the nucleus. This leads to mitogenic progression, suggesting that patched-1 may have cell cycle gatekeeper functions.
Mutations in the gene encoding the p53 tumor suppressor have been found in more than half of sporadic BCCs. The inactivating mutations usually bear evidence of UV induction, bearing CC → TT and C → T substitutions produced by the photoproducts of adjacent pyrimidines. In one study of the prevalence of TP53 and PTCH1 mutations in sporadic BCCs, it was found that among 18 BCCs, 61% demonstrated loss of 9q markers ( PTCH1 ), 61% had acquired TP53 mutations, and 39% had alterations in both genes. In subsequent studies, 38% of early onset BCCs were found to have mutations in both TP53 and PTCH1 and 75% of all BCCs were found to have allelic loss of 9q and a TP53 mutation.
In addition to BCNS, non-syndromic multiple basal cell carcinomas (OMIM 605462) have been described in several families, one in which male-to-male transmission was noted and three which were strictly unilateral. Whether these presentations represented mutations in genes known to be associated with BCC in a mosaic pattern has not been determined. More recently, a family was described in which numerous BCCs developed in a generalized distribution, most likely in an autosomal dominant fashion.
In Rombo syndrome (OMIM 180730), multiple basal cell carcinomas are accompanied by vermiculate atrophoderma, milia, hypotrichosis, telangiectasias and acral erythema. Bazex syndrome (Bazex–Dupre–Christol syndrome, OMIM 301845) is characterized by the triad of multiple basal cell carcinomas, congenital hypotrichosis, and follicular atrophoderma. Whereas the gene defects which give rise to these syndromes have yet to be determined, the X-linked mode of inheritance of Bazex syndrome suggests that additional genes within the same pathway or novel pathways have yet to be determined.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here