Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Pain is associated with considerable variability between individuals. Humans exhibit robust differences in their thresholds and tolerances to controlled noxious stimuli, in their analgesic response to drugs, and in their susceptibility to (and severity of) clinical pain syndromes. In fact, the central focus of pain research can be accurately cast as a question of individual differences: why does chronic pain eventually develop in only a minority of people after injuries and infections that can produce chronic pain (e.g., traumatic nerve injury, stroke, herpes zoster, diabetes)? As with all biological phenomena, this variability is produced by some combination of mostly undetermined genetic and environmental factors. Although it is likely that environmental factors account for more of the variance overall, the study of genetic factors associated with trait variability boasts the advantage that although the number of possibly relevant genes is large (>20,000), it is ultimately quite finite. Methodological advances have rendered the genetics of pain tractable for study in laboratory animals and humans, and genes are being identified in both species at an increasing rate. The present chapter addresses the current methods and findings of pain genetics in mice and humans. Success in these endeavors promises not only to identify novel pain-related molecules and advance our understanding of pain pathophysiology but perhaps also to eventually allow individualized prediction and treatment of pain.
† Deceased.
The robust interindividual variability observed in sensitivity to pain, the propensity for chronic pain conditions to develop, and the response to analgesic manipulations is a scientific and clinical challenge. Laboratory studies have documented impressive individual differences in thresholds, tolerance, and pain scale ratings of noxious experimental stimuli ( , , ). Impressive correlations between pain ratings and simultaneously obtained measures of cortical activation with functional magnetic resonance imaging or positron emission tomography ( , ) suggest that these individual differences truly reflect variable perception and not variable scale utilization. Epidemiological studies of chronic pain syndromes known to develop after specific traumatic or infectious insults consistently reveal that chronic pain will eventually develop in only a small fraction of patients ( , , ). Thus, these insults are not themselves sufficient to produce chronic pain; some factor intrinsic to the recipient of the injury is also to blame. The explanation is very likely to be a classic example of genetic–environmental interaction: both the injury and some innate or acquired propensity are necessary. A potentially useful fact is that individual differences in laboratory pain sensitivity are predictive of clinical pain severity and response to treatment ( ).
Impressive interindividual variability has been documented in response to experimental and clinical administration of analgesics as well, including opioids ( , , , ), placebo ( , , ), and non-steroidal anti-inflammatory drugs ( , , ).
An understanding of the basis of such individual differences would have a number of benefits for therapeutic development efforts, prediction of risk, and individual tailoring of existing therapies. Obviously, environmental factors are important in the final explanation, both in terms of their unique additive effects and in their interaction with pain-relevant genetic factors. This review, however, aims to describe only techniques and recent progress in pain and the genetics of analgesia in laboratory animals and humans.
In theory, many genetic approaches are as tractable in humans as in laboratory animals. For example, identification of trait-relevant genes by linkage mapping or association study can easily be performed in humans. Why then bother studying pain genetics in laboratory rats and mice? One reason is the statistical power and experimental simplicity afforded by controlled crosses, which obviously can be attempted only in laboratory animals. Starting with inbred progenitors (see below), a single set of grandparental breeders can easily beget hundreds of the genetically segregating F 2 hybrid or backcross mice needed for mapping complex traits by linkage. Simultaneous evaluation of the many existing inbred strains themselves can now also be used for gene identification by haplotype mapping. By contrast, even the largest human pedigrees are modest in size and genetically complicated (with more than two possible alleles at each locus) in comparison. Single-gene association studies in humans remain poorly replicable ( ), and the powerful method of full-genome, single nucleotide polymorphism (SNP)–based linkage disequilibrium mapping (i.e., genome-wide association studies [GWASs]) has not quite yet been applied to pain, at least not in the peer-reviewed literature.
If we are to use laboratory animals to study pain genetics, we must ask to what extent are data derived from such animals relevant to our species. There is no particular reason to believe that specific DNA sequence variants common in Mus musculus , say, would be preserved in human beings, the lineages of the two species having diverged more than 100 million years ago. However, there is ample reason to expect that genes relevant to pain in mice would also be relevant to pain in humans. On completion of the human and mouse genome sequences, performed a detailed analysis of the genes on mouse chromosome 16. Of the 731 predicted genes, only 14 (1.9%) had no homologues in the human genome. The evolutionarily ancient role of nociception suggests that mammals ought to be quite similar in genetic and physiological processing of this particular biological trait, even though humans probably have a more sophisticated cognitive and emotional dimension to their pain.
Although any number of rat strain differences of relevance to pain have been demonstrated (see , , , , , , , , ), the mouse has clearly become the default laboratory subject for genetic research. One reason is the much larger number of commercially available inbred mouse strains ( ) than inbred rat strains. Another is the relatively more dense genetic map in the mouse, which led to completed sequencing of the mouse genome ( ) several years before that of the rat ( ). Probably the most important reason for ascendance of the mouse in pain genetics research was the unique ability in this species to create transgenic knockout mice (although rat knockouts can now be constructed; ), which caused intense interest in the normative responses of the inbred mouse strains in which such mutations are created.
There are two related problems to which pain genetics is addressed. One type of pain genetics asks the question, which genes are relevant to pain? This is merely a restatement of the fundamental question of reductionist pain biology, which aims to define the molecular “players”: the proteins playing a role in mediation of pain perception and modulation. There are considerable advantages to identifying pain-relevant proteins by identifying the genes coding for them in that genetic approaches offer simplicity (there are only ~30,000 genes but many more proteins) and unparalleled specificity (typically, DNA or mRNA sequences of >15 nucleotides unambiguously define one and only one gene). The second type of pain genetics asks the question, of the pain-relevant genes, which are responsible for individual differences in sensitivity to pain and analgesia and for differential susceptibility and/or expression of painful pathologies? This second question represents classic, mendelian genetics as applied to pain traits. Along the way to answering these questions, one can adopt either a “bottom-up” (genotype → phenotype) or a “top-down” (phenotype → genotype) strategy ( Fig. 10-1 ). With a bottom-up strategy, one focuses on a particular gene (usually identified via the known role of its protein product in pain) and studies the relationship between expression of that gene and some systems-level pain phenomenon. That is, one can measure or alter expression of the mRNA transcripts of individual genes to provide evidence of involvement of these genes—and thus their proteins—in some aspect of pain physiology. With a top-down strategy, one examines populations showing contrasting systems-level phenotypes (e.g., different strains, nerve-damaged versus intact animals) and tries to find the genes responsible for the differences. The responsible genes will produce either proteins with a change or deletion of an amino acid or acids in the populations being compared or proteins that are alternatively spliced to reflect different exons. In other cases, the gene variants might not affect protein structure at all but rather the basal expression levels of these proteins. The various methods in current use are discussed briefly; their respective advantages and disadvantages in the study of pain genetics have been addressed previously ( ).
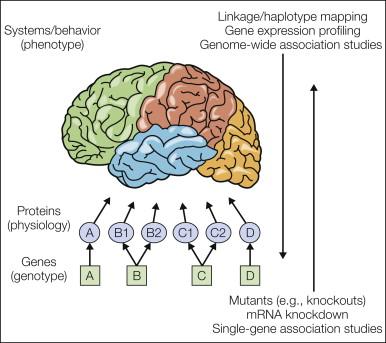
By far the most common bottom-up approach to pain genetics is the creation and testing of transgenic knockout mice. These are genetically engineered null mutants in which a single gene is effectively ablated via homologous recombination of embryonic stem cells with a transgenic targeting vector (see ). Such knockout mice are compared with “wild-type” and heterozygous littermates, and if the genetic “lesion” results in an aberrant behavioral and/or biochemical pain phenotype (e.g., altered sensitivity on an algesiometric assay), this demonstration provides evidence, subject to certain caveats (see ), that the gene is required for normal expression of the trait. A large number of knockout mice have been tested for nociceptive and analgesic sensitivity; we maintain a web database of findings from such experiments called the Pain Genes Database ( ). As of this writing, the pain-related phenotypes of 322 mutants have been documented (see http://paingeneticslab.ca/ 4105/06_02_pain_genetics_database.asp). A comprehensive review of transgenic studies of pain is beyond the scope of this chapter, however, and findings using this technique will appear elsewhere in this volume. To give the reader a sense, Table 10-1 lists simply the number of genes currently demonstrated to affect pain, hypersensitivity after inflammatory and neuropathic injury, and morphine analgesia via the transgenic technique. It should also be noted that spontaneously occurring mutations (of coat color genes and genes causing obvious phenotypic abnormalities) have been and continue to be studied for their relevance to pain (see ; also see for review). Great sums of money have been spent on the deliberate induction, via chemical mutagenesis, of random point mutations combined with systematic screening for phenotypic alterations. Three public mutagenesis projects ( , , ) included a nociceptive assay (hot plate test), but data describing only one pain-relevant mutant were ever published ( ). A genome-wide screen of Drosophila mutations recently identified a thermal avoidance gene in flies that appears to play a similar role in humans ( ) (see below).
| TRAIT | ASSAY/ DRUG/ETIOLOGY | INCREASED ∗ | DECREASED † |
|---|---|---|---|
| Analgesia | Morphine | 25 | 28 |
| Cold allodynia | Neuropathic | 2 | 14 |
| Heat hyperalgesia | Inflammatory | 17 | 47 |
| Neuropathic | 5 | 35 | |
| Inflammatory pain | Formalin (early phase) | 14 | 37 |
| Formalin (late phase) | 20 | 74 | |
| Abdominal constriction | 9 | 34 | |
| Capsaicin | 5 | 28 | |
| Mechanical allodynia | Inflammatory | 19 | 58 |
| Neuropathic | 13 | 74 | |
| Mechanical pain | Paw/tail pressure | 7 | 19 |
| von Frey | 17 | 30 | |
| Thermal pain | Hot plate | 36 | 61 |
| Paw withdrawal | 15 | 29 | |
| Tail flick/withdrawal | 25 | 41 |
∗ Null mutant (knockout) mouse significantly more sensitive than wild-type mouse.
† Null mutant (knockout) mouse significantly less sensitive than wild-type mouse.
In the original version of this bottom-up approach, called antisense knockdown, an oligonucleotide is synthesized to be complementary (i.e., in an antisense orientation) to an mRNA transcript of the gene of interest and injected into a target tissue. By a mechanism involving steric hindrance in ribosomes and/or enzymatic degradation, the antisense oligonucleotide prevents (to some degree, hence “knockdown” instead of “knockout”) successful translation of the native mRNA into protein ( ). Although the antisense approach has several major advantages over knockout mice (see ), it is technically difficult and has been used fairly sparsely in pain research. When successful, however, it has provided impressive demonstrations of the role of particular genes in pain (e.g., , , ).
More recently, mRNA knockdown in vitro and in vivo has been achieved with the use of small interfering RNA (siRNA; see , ), which provides much greater translational block efficiency. The role of any number of genes in pain has been demonstrated via in vivo knockdown using siRNA, known as RNA interference ( ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ).
Selective breeding is the oldest of the top-down approaches and has been performed in agriculture and animal husbandry for millennia. Also known as artificial selection, the technique as applied scientifically involves two-way breeding of a genetically heterogeneous stock of animals based on their response in regard to a trait of interest (see ). Only extreme responders are used to beget the next generation of offspring, and the resultant “high” and “low” lines will start to diverge phenotypically for all heritable traits. The great advantage of the technique is that all trait-relevant genes will eventually become fixed in alternate allelic states in the two lines such that very robust phenotypic differences are demonstrated. Of greatest relevance to pain genetics are three selective breeding projects: the HA/LA rat lines selected for high and low autotomy after nerve transection ( ), the HA/LA mouse lines selected for high and low analgesia after forced swim stress ( ), and the HAR/LAR mouse lines selected for high and low analgesic response to levorphanol (and in later generations, morphine) ( ). Much has been learned from these lines (see ); however, it remains difficult to identify the fixed genes, and this approach is rarely used today.
The use of inbred strains for analysis of classic pain genetics is the dominant modern top-down strategy. Inbred strains are produced by repeated brother × sister mating over at least 20 generations, a breeding scheme that eliminates all genetic heterozygosity and renders individual members of the strain isogenic (i.e., clones) to all others, barring the very rare occurrence of new mutations ( ). Many existing inbred mouse strains are the descendents of European and East Asian “fancy mice” bred at the turn of the 20th century by Abbie Lathrop and William Castle, and excellent genealogical records are available in many cases ( ). A comparison of randomly selected inbred strains allows one to survey alleles of trait-relevant genes existing in the European and East Asian “fancy mouse” population of the early 1900s fixed into a homozygous state in every strain but into a different state in phenotypically contrasting strains. That is, each laboratory inbred strain is a different fixed “mosaic” of M. musculus domesticus , M. musculus castaneus , and M. musculus musculus alleles ( ). Inbred strains derived from stocks of Japanese wild mice ( M. musculus molossinus ) may have even greater genetic variability ( ). Because they are isogenic, inbred strains are tremendously useful for establishing the heritability of traits since within-strain variability is by definition of environmental origin and between-strain variability is very probably genetic. An inbred “strain survey” can identify the most robust pair-wise strain differences, and these extreme-responding strains are the ideal progenitors of linkage-mapping populations.
Linkage mapping, which when applied to complex traits is known as quantitative trait locus (QTL) mapping, searches for genomic regions that are co-inherited with trait variability in genetically segregating populations (e.g., F 2 intercross, backcross, recombinant inbred strains) (see ). As a practical matter, one correlates the phenotypic responses of, say, F 2 hybrid mice with their inherited genotype at non-coding, polymorphic DNA markers (microsatellites or SNPs), and where such genetic “linkage” is established then infers the existence of a trait-relevant gene or genes in the genomic vicinity of the markers. By contrast to all other strategies in current use, QTL mapping allows one to identify genes containing the very polymorphisms that are the cause of the original strain difference since what is being examined is not gene expression but the DNA sequence itself. The disadvantage of QTL mapping is that identifying the broad genomic region linked to the trait (i.e., the QTL) is far easier than “positional cloning,” or identifying the gene and polymorphism responsible ( ). Positional cloning has historically been attempted by creating advanced mapping populations such as congenic strains ( ), followed by cloning and sequencing.
A very useful advance is haplotype mapping, also known as in silico QTL mapping ( ; also see ). Because DNA variants are not inherited independently but rather in “blocks” known as haplotypes (see later) and since sequencing in multiple inbred mouse strains has already occurred, one can now simply search for statistical associations between strain means and haplotypic patterns. With a sufficiently high number of strains, significant linkages can be evinced, and the mapping resolution is extremely good (generally within the genes themselves or very close to them). Despite many false positives, this technique has been used profitably in mouse pain genetics in recent years.
The most recently developed top-down strategy is microarray or “gene chip” expression profiling. The technique, developed by Pat Brown’s laboratory at Stanford ( ) and made widely available by commercial entities such as Affymetrix, Inc., allows quantification of mRNA expression in a massively parallel fashion (see ). Other related techniques, such as differential display polymerase chain reaction ( ) and massively parallel signature sequencing ( ), allow the same sort of analysis. To run a profiling experiment, one isolates the total RNA from at least two contrasting tissue sources (e.g., dorsal root ganglia from a rat in neuropathic pain versus dorsal root ganglia from a control rat), and after a series of reverse transcription, amplification, and labeling steps, hybridizes the samples to a nylon or glass array. In the common Affymetrix version, the glass “chip” contains synthesized oligonucleotides corresponding to many thousands of known and unknown gene transcripts in catalogued locations. The fluorescent probes are excited by a laser and detected with a scanning confocal microscope; the intensity and color of fluorescent labeling at each location correspond to the absolute and relative (between groups) abundance of that particular gene’s mRNA in the tissue.
To date, approximately 30 microarray studies directly focused on chronic pain and using standard assays have been published ( ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ; ). The results of all these investigations (also see , , ) illustrate the power of and problems with the approach. Depending on the criteria used to establish “significance,” these studies have identified up to hundreds of genes up- or down-regulated by the injury. On the plus side, many of these genes were not previously known to play any role whatsoever in chronic pain, and thus the heuristic potential is immense. Most investigators were surprised, for example, by the large number of “hits” among genes relevant to neuroinflammation ( , ). Various forms of cluster analysis applied to microarray data may identify groups of genes that are co-regulated ( ), an impossible task with other approaches. However, it is difficult to evaluate the relative importance (and cause-and-effect relationships) of these hundreds of genes and also very difficult to determine which of them are specifically related to pain versus the nerve injury itself. It is also difficult (without confirmation by other techniques) to separate true from false positives. We recently performed a meta-analysis of the first 20 published chronic pain–relevant microarray studies ( ).
As mentioned, the major aim of this chapter is to review the genetics of individual differences since the molecular genetics of pain is addressed elsewhere in this volume. Therefore, in this final section we discuss findings of two types: (1) general principles of pain genetics derived largely from selective breeding and inbred strain comparisons and (2) results of pain-relevant QTL and haplotype mapping studies. Interested readers are directed to other reviews as well ( ; ; ).
Heritability ( h 2 ; see later for further discussion) is the proportion of trait variance attributable to genetic inheritance. Heritability can be estimated in laboratory animals either by using panels of inbred strains or by assessing the response to artificial selection (see ). A large number of studies noting pain-relevant strain differences have compared the thermal nociception–sensitive and opioid analgesia–resistant C57BL/6 mouse strain with the nociception-resistant and opioid analgesia–sensitive DBA/2 strain (see , ). These strains do not represent the extreme responders among commonly available mouse strains, however, and in any case pair-wise comparisons do not provide much power to estimate heritability. In the only systematic inbred strain surveys to date, the heritability of response to 22 common algesiometric assays has been found to range from h 2 = 0.30–0.76, with a median value of 0.46 ( , ). This range of heritability is highly similar to that obtained in large human twin studies of experimental pain published in recent years ( , ). Using the same set of strains, the heritability of the efficacy of eight analgesics was found to be slightly lower, h 2 = 0.12–0.45, with a median value of 0.29 ( ). The three existing selective breeding studies of relevance to pain (see earlier) have yielded heritability estimates of h 2 = 0.30 for autotomy ( ), h 2 = 0.32 for levorphanol analgesia ( ), and h 2 > 0.32 for swim stress–induced analgesia ( ).
One interesting use of both selected lines and inbred strains involves the investigation of genetic correlations (see ). Most genes are known or thought to be pleiotropic, that is, affecting more than one trait. If selection or chance has fixed the alleles of a gene into a homozygous state, that fixation will probably affect multiple traits. Genetic correlation is demonstrated if lines selected for their performance on one trait are observed to differ on another or if the same distribution of phenotypic responses on two traits is observed among a panel of inbred strains.
As might be expected, the sensitivity of inbred strains and selected lines on certain nociceptive assays is genetically correlated with sensitivity on other nociceptive assays, but not all other nociceptive assays. Inbred mouse strains sensitive to tail flick test nociception are by and large the same mouse strains sensitive to paw withdrawal (Hargreaves’) test nociception. By contrast, a largely different set of strains are sensitive to formalin test nociception ( ). In our analysis of 22 nociceptive assays in a common set of 12 strains, we provided evidence using multivariate analysis of five “clusters” of assays: (1) thermal nociception, (2) ongoing nociception from chemical stimuli, (3) mechanical hypersensitivity, (4) thermal hypersensitivity, and (5) afferent-dependent (featuring initial ongoing nociception) thermal hypersensitivity ( ). Since variability measured on tests within a cluster should be mediated by common genes but variability on tests in different clusters by different genes, we believe that these clusters represent fundamental “types” of pain in the mouse. This is a nice example of how a genetic approach can address a non-genetic question. Species specificity may apply, however, since similar studies using rat strains have yielded somewhat different results ( , ).
Another remarkable genetic correlation pertains to analgesia of different modalities. Such correlation was hinted at by findings in selected lines, for example, when mice selected for high and low stress-induced analgesia were found to be high and low responders, selectively, to levorphanol, morphine, and the κ-opioid agonist U50,488 ( , ). We recently evaluated the analgesic potency of five neurochemically distinct analgesics in 12 inbred strains: morphine; U50,488; clonidine; epibatidine; and the cannabinoid agonist WIN55,212-2 ( ). Although these drugs all activate descending pain-inhibitory mechanisms in the central nervous system, they bind to five distinct molecular sites. Thus, we were surprised to discover a remarkably high degree of genetic correlation among them ( r = 0.33–0.68). This finding has important implications. First, it appears that a “master” analgesia variability gene or set of genes must exist, and it is highly unlikely that this gene is related to the binding site of each drug. If this is true, current attempts to explain variable analgesic drug action (i.e., pharmacogenetics) solely by searching for polymorphisms in the binding site gene will be incomplete. In a follow-up study, a similar genetic correlation was observed between acetylsalicylic acid and indomethacin analgesia, but acetaminophen analgesia appeared to be genetically distinct ( ).
A related genetic correlation is the repeatedly noted correlation between baseline nociceptive sensitivity and subsequent sensitivity to inhibition of the stimulus by analgesics ( , , ). That is, mouse strains initially sensitive to nociception are also resistant to analgesia, and vice versa. This principle works within a number of nociceptive modalities and suggests that the “master” analgesia gene or genes referred to above may in fact be ones with a primary action on nociception, which may subsequently affect analgesia as well. hypothesized that this can be explained in terms of genetic differences in effective stimulus intensity affecting fractional receptor occupancy of the analgesic.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here