Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
In 1953, Watson and Crick published their landmark paper on the molecular structure of nucleic acids and a particularly prescient comment was made: “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.”
The pace of technological innovation and our resulting understanding of the relationship of genetics to human disease continues to increase exponentially. Today, the ability to partially or completely sequence the genetic code of an individual is an inexpensive commodity, and the current and future impact of this knowledge continues to increase.
With this knowledge, our understanding of disease associations with the genetic code has become more complex, beyond simple assessment of various types of mutations leading to a certain phenotype. This is driven by an ever-increasing understanding of the interplay between genetic and environmental factors in disease. Today, in addition to easily understanding environmental factors important in cardiovascular disease (e.g., smoking, diet, exercise, and many others), it is apparent that the environment interacts with the genome in complex ways. Epigenetics, which is the study of changes in gene modification, or translation-altering gene expression, is now understood to be a mechanistic explanation, and possibly a therapeutic target for these environmental–genetic interactions ( Fig. 3.1 ). One classic example of epigenetics in clinical medicine was the observation of children born to famished mothers during the Dutch Hunger Winter of 1944 to 1945. These children were observed in their sixth decade of life and were found to have higher rates of obesity and coronary artery disease (CAD). Further evaluation showed them to have decreased methylation of insulin-like growth factor-2, a factor known to be important in metabolism, energy use, and weight, compared with their genetically similar siblings. The increased understanding of factors that alter methylation, histone packaging, and noncoding RNA, to name a few, will lead to a stronger grasp of the correlation between disease and the genetic code. Because of the growing complexity and rapid pace of new information presented, the development of genetic specialists within each field has been paramount to assist healthcare providers in deciphering this information and its clinical implications.
The goal of this chapter is to introduce the clinically important principles of genetics and the application of these principles to clinical medicine, with particular emphasis on the genetics of cardiovascular diseases ( Table 3.1 ). A brief glossary of the clinically important terms in this chapter is shown in Box 3.1 .
| Phenotype | Genetic Alteration (Most Common) | Functional Effect of Mutation | Method of Inheritance | Prevalence | Disease Characteristic |
|---|---|---|---|---|---|
| Cardiomyopathy | |||||
| HCM a | MYPBPC3, TNNI3, TNNT2, TPM1, MYL3 | Sarcomere protein mutation leading to myocardial disarray | Autosomal dominant | 0.02%–0.23% | LVH, LVOT obstruction, ventricular tachycardia |
| Fabry disease | GAL-A (GLA) | Loss of function of α-galactosidase-A in lysosomes, leading to build up of globotriaosylceramide in organs, including the heart | X-linked | 0.0025% | LVH, systolic and diastolic dysfunction, coronary artery disease, conduction abnormalities, valvular dysfunction |
| ARVD | JUP, DSP, PKP2, DSMG2, DSC2, TGFβ3, TMEM43 | Desmosome dysfunction leading to loss of electrical coupling between myocytes, leading to cell death with fibrofatty replacement and arrhythmias | Autosomal dominant | 0.02%–0.05% | Fibrofatty replacement of RV myocytes. RV regional (early) or diffuse (late) disease. LV abnormalities can exist. Depolarization and repolarization abnormality leading to ventricular tachycardia and fibrillation |
| Familial DCM a | LMNA, MYH6, MYH7, MYPN, TNNT2, SCN5A, MYBPC3, etc. | Predominantly genes affecting stability of inner nuclear membrane, sarcomeric protein, or ion flux, though not exclusively | Variable | 20%–35% of patients with idiopathic DCM | LV or biventricular dysfunction |
| Arrhythmia | |||||
| LQTS a | Autosomal dominant | 0.014%–0.04% | Prolonged corrected QT >440 ms. Propensity VT/VF and SCD | ||
| Type 1 | KCNQ1/KVLQT1 | Mutation in α subunit of I Ks | |||
| Type 2 | KCNH2/HERG | Mutation in α subunit of I Kr | |||
| Type 3 | SCN5A | Mutation in α subunit of I NA | |||
| Brugada syndrome a | SCN5A, GPD1-L CACNA1c |
Mutation in α subunit of I NA Mutation in α subunit of I CA |
Autosomal dominant | Abnormal ECG with RBBB with characteristic cove-shaped ST elevation in leads V 1 –V 3 without secondary causes. Propensity for VT/VF and SCD | |
| Coronary Artery Disease | |||||
| Autosomal dominant familial hypercholesterolemia a | LDLR, APOB, PCSK9 | Inability for LDL-R to LDL-C binding lead to diminished catabolism of LDL-C leading to elevated plasma levels of cholesterol | Autosomal dominant | Elevated LDL subparticles lead to accelerated atherosclerosis, leading to coronary artery disease | |
a Denotes only a select example of known genetic abnormalities.
Alleles: Copies of a specific gene. Humans have two alleles for each gene (one each from the biological father and mother). Alleles may have functional differences in their DNA sequence. A person with two identical copies of an allele is termed homozygous; a person with two different copies of an allele is called heterozygous.
Dominant mutation: A mutation in one allele of a gene that is sufficient to cause disease. More severe disease or lethality may result from a dominant mutation in both alleles of a gene.
Environmental effects: For this chapter, any potentially controllable influence on an individual. Examples are diet, exercise, air quality, a response to a prescribed or an over-the-counter medication, cigarette smoking, and alcohol use.
Genotype: The genetic makeup of an individual. Genotype can refer to specific genes or to the overall genetic profile.
Mutation: Changes in the DNA sequence of a gene that result in a gene product (protein) that has an altered sequence. For this chapter, mutations are considered to be changes in the DNA sequence of a gene that result in either loss of function or severely altered function.
Phenotype: The functional effects of genetic changes together with environmental influences. For instance, a person's appearance (body build, muscularity, hair color), the presence of measurable abnormalities that may reflect underlying disease processes, or other physical features demonstrate phenotypes. The list of measurable abnormalities is almost infinite, from blood pressure abnormalities to abnormal biochemical measurements (e.g., serum glucose levels) to ECG abnormalities reflecting ion channel abnormalities (as occur in LQTS) to coronary heart disease measured by angiography or endothelial dysfunction measured by forearm blood flow variability.
Polymorphism: An inherited variation in the DNA sequence of a gene that occurs at a greater frequency than would be expected of a mutation. Humans have thousands of polymorphisms, none of which are believed to be solely responsible for disease. Technically no different from mutations (a change in DNA sequence from “normal”), polymorphisms typically alter the gene product more subtly than mutations. It is believed that many human phenotypes result from the interplay between an individual's mix of polymorphisms and the environment.
Recessive mutation: A mutation that requires alterations in both alleles of a gene to cause disease (except in the case of mutations in the X and Y chromosomes).
LQTS , Long QT syndrome.
In the coming years, it is likely that genetic analysis will play an increasingly important role in understanding high-risk cardiovascular syndromes. Significant advances have been made in several cardiovascular phenotypes, as illustrated by the examples in this chapter.
Cardiomyopathy is a term that refers to a heterogeneous group of diseases that affect the myocardium. Cardiomyopathies result from multiple etiologies, including ischemia, nonischemic causes (restrictive or infiltrative), and hypertrophy, each of which has subcategories. The study of familial trends and transmissible genetic abnormalities in cardiomyopathies has led to more detailed classification based not only on phenotypic appearance, but also on genetic alterations. In some cases, this knowledge is of importance for risk stratification and treatment.
A classic example is hypertrophic cardiomyopathy (HCM), which is defined as the presence of myocardial hypertrophy caused by myocardial disarray. HCM is a disease that affects 0.02% to 0.23% of the general population, and in some, but not all HCM patients, it results in symptomatic diseases due to left ventricular outflow obstruction and an increased risk for fatal arrhythmias, such as ventricular tachycardia. Current understanding of the genetic basis of HCM is complex ( Fig. 3.2 ). Most cases are caused by autosomal dominant mutations in genes that encode proteins in the sarcomere. Beta-myosin heavy chain and myosin-binding protein C are the most common genes affected; however, other genes in the sarcomere protein, including troponins I and T, the tropomyosin α-1 chain, and myosin light chain-3 have also been implicated in HCM.
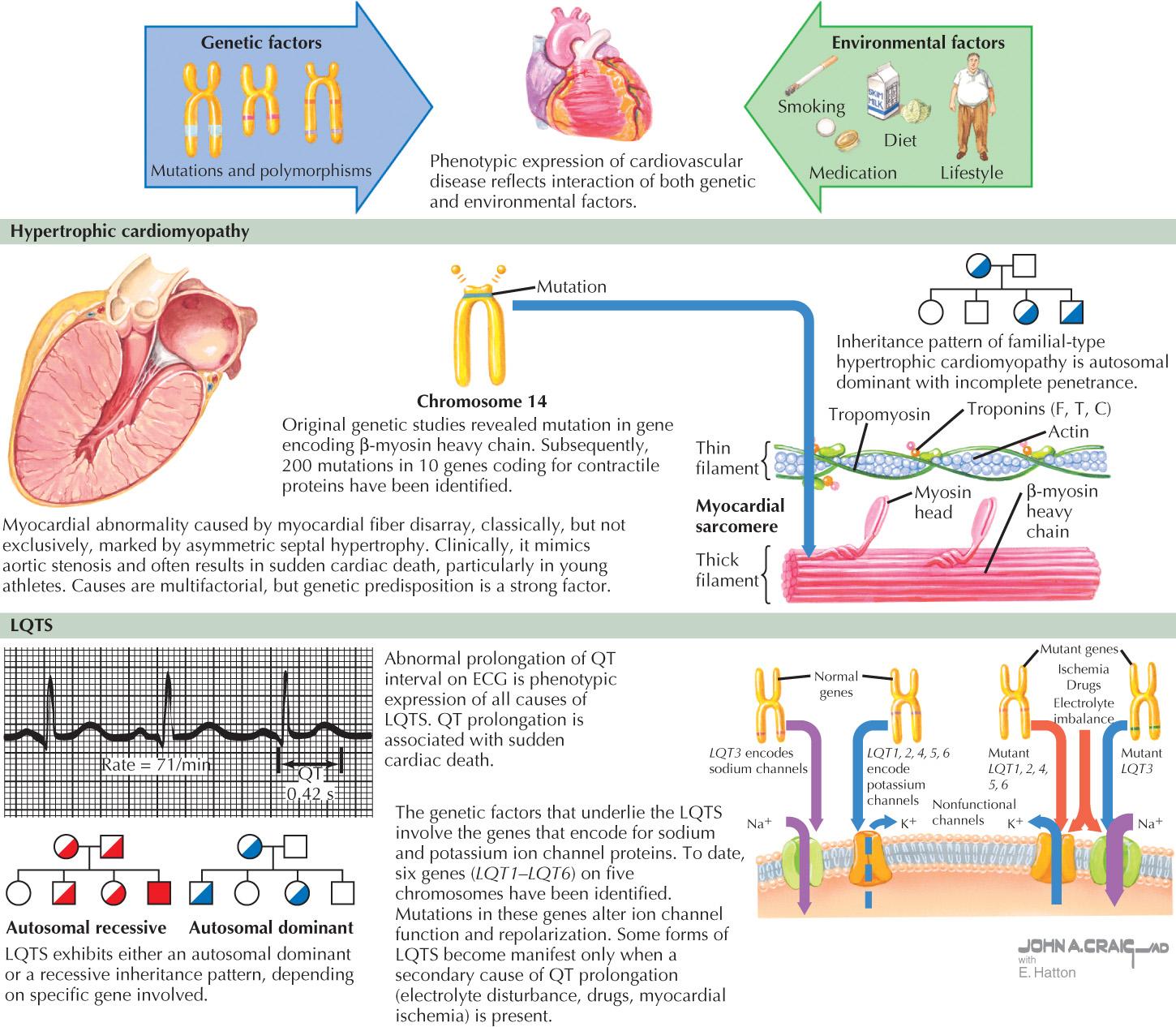
The specific genetic basis for HCM in a given family has allowed a much better understanding of the disease and its management.
For those with an identifiable genetic cause of HCM, genetic screening of family members allows for more focused disease surveillance and management at an early stage before symptoms develop. For those in the same family who lack the causative mutation, surveillance can be less frequent, or in some cases, not necessary. The treatment of left ventricular outflow obstruction is not driven by genetic mutations per se; however, combining pedigree–phenotype analysis with DNA sequence information can identify those with an increased risk of sudden cardiac death (SCD) and may be used for decision making in the use of implantable cardioverter-defibrillator. In addition, genetic testing can identify a subset of individuals with a positive genotype but a negative phenotype, which raises new clinical questions, such as whether these individuals should be excluded from participation of competitive sports, would benefit from a prophylactic defibrillator, and how often they should be screened for phenotypic HCM.
Genetic etiologies also underlie numerous autosomal recessive and X-linked metabolic causes of cardiomyopathy, such as Anderson-Fabry disease, mitochondrial cardiomyopathies, neuromuscular diseases, and infiltrative diseases. Certain drugs, such as anabolic steroids and tacrolimus, are also therapies that have been shown to be causes of HCMs.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here