Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
In 1921 James Ewing described the cancer that came to carry his name: a primary bone tumor composed of small round blue cells and devoid of the malignant osteoid that characterizes osteosarcoma. Subsequently, pathologists described other clinicopathologic entities initially thought to be distinct, such as peripheral primitive neuroectodermal tumor (PNET) of bone or soft tissue or the Askin tumor of the chest wall. However, biologic study and clinical responses to chemotherapy have now linked these tumors into one entity or family of tumors, a grouping that also includes extraosseous Ewing sarcoma. Commonly used terms for this tumor include Ewing sarcoma, Ewing tumors, and Ewing sarcoma family of tumors. In this chapter the World Health Organization–approved term, Ewing sarcoma, is generally used. When appropriate, specific characteristics that differentiate Ewing sarcoma, PNET, and Askin tumors are described.
The chapter begins with a discussion of the epidemiology of Ewing sarcoma. Next, the current understanding of the cellular and molecular features of these tumors is reviewed. An overview of the clinical presentation and prognostic features of patients with this disease are presented. This overview is followed by a discussion of the management of patients with Ewing sarcoma. The chapter concludes with a review of the late effects seen in patients treated for these tumors.
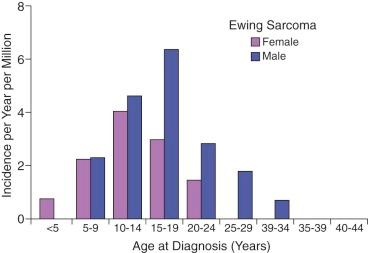
Ewing sarcoma is the second most common type of primary bone cancer in the United States and Europe, accounting for approximately 25% to 34% of malignant bone tumors. Approximately 250 new cases of Ewing sarcoma are diagnosed in the United States each year. In the United States Ewing sarcoma has had a relatively stable average incidence of 2.5 to 3 cases per million per year from 1975 to 2000. A similar age-adjusted incidence has been noted in Europe. As shown in Figure 61-1 , the peak incidence occurs during adolescence, with an average incidence of 4.6 cases per million per year for the 15- to 19-year age range. Younger children are affected less frequently, and the disease is distinctly uncommon in adults older than 35 years of age. A male predominance has been noted, with a male-to-female ratio of approximately 1.25 : 1. However, one study reported a female predominance in patients younger than 3 years of age at initial diagnosis. There is no seasonal variation in the appearance of this disease.
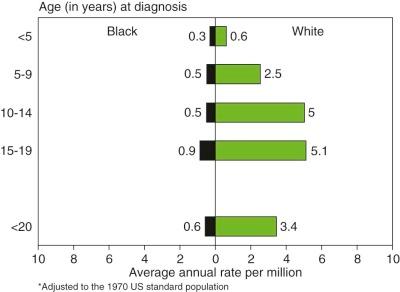
The incidence of Ewing sarcoma varies markedly among races and countries, with the highest frequency reported from Australia and Sao Paolo, Brazil. People of African ancestry appear to have the lowest incidence of this disease. Figure 61-2 demonstrates the dramatically lower incidence of these tumors in the U.S. African-American/Black population. A similar low incidence in sub-Saharan Africa has been reported and suggests a possible genetic difference in the risk of developing Ewing sarcoma. Patients of African ancestry with Ewing sarcoma have higher rates of soft tissue tumors and an older age distribution compared with patients of European ancestry, indicating that racial differences in this disease extend beyond differences in incidence. Ewing sarcoma also appears to be uncommon in people of East Asian ancestry, based on data from China, Japan, and Thailand. Data from the Middle East, North Africa, and the Indian subcontinent suggest that Ewing sarcoma is at least as common in these populations as in people of European ancestry. In small studies from Kuwait and Bombay, for example, Ewing sarcoma was identified as the most common primary malignant bone tumor.
Based on these geographic differences in the incidence of Ewing sarcoma, several groups have investigated the genetic epidemiology of this disease. One group reported differences between African and European populations in the number of Alu repeat sequences in intron 6 of the EWSR1 gene, upstream of the breakpoint region involved in translocations at this locus seen in most cases of Ewing sarcoma. Two studies further focused on this possibility that differences in the incidence of Ewing sarcoma may be due to differences in polymorphisms in the EWSR1 locus that could result in different rates of translocation. Both studies each identified a different single nucleotide polymorphism with low-level statistical association with Ewing sarcoma, although the effect of each polymorphism on the risk of Ewing sarcoma was small. Another study investigated the incidence of other tumors that harbor EWSR1 translocations across racial groups and observed higher rates of desmoplastic small round cell tumor in the U.S. African-American population compared with the U.S. European-American population. This finding of higher rates of an EWSR1 -associated tumor in a population with a lower rate of Ewing sarcoma provides additional evidence that differential susceptibility to translocation at the EWSR1 locus does not account for racial differences in the incidence of Ewing sarcoma. More recently, a genome-wide association study identified and validated polymorphisms in three loci, 1p36, 10q21, and 15q15, associated with increased risk of Ewing sarcoma. These risk variants are present at lower frequencies in African populations, providing one potential explanation for the lower rates of Ewing sarcoma in this population.
The etiology of Ewing sarcoma remains largely obscure. These tumors are not generally believed to be familial, although rare cases of sibling pairs with Ewing sarcoma have been reported. First- and second-degree relatives of patients with Ewing sarcoma do not appear to have an increased overall risk of cancer, although a higher rate of stomach cancer, melanoma, renal cancer, and brain tumors has been reported.
Patients with Ewing sarcoma appear to have an increased incidence of specific congenital anomalies. The most consistent finding has been an association between Ewing sarcoma and congenital hernias, particularly inguinal hernias. A pooled analysis of the available literature has confirmed this association, with an odds ratio of developing Ewing sarcoma of 2.8 for children with congenital hernias compared with children without congenital hernias. Whether a common hormonal or environmental mechanism underlies the development of both congenital hernias and Ewing sarcoma remains unclear. Patients with Ewing sarcoma have also been noted to have an increased incidence of bone anomalies, particularly rib and vertebral anomalies. Other congenital anomalies, including cataracts and genitourinary anomalies, have been reported in patients with Ewing sarcoma. These anomalies occur at a low incidence in these patients and without a consistent pattern, suggesting that their occurrence with Ewing sarcoma may be coincidental.
Various parental exposures have been associated with the risk of developing Ewing sarcoma. The most consistent finding has been an association between parental farm exposure around the time of conception through birth and the subsequent development of Ewing sarcoma. This association has been reported in a series of case-control studies and affirmed in a meta-analysis, with an increased risk if either parent was involved in farming. Specific exposures related to agricultural work that have been suggested as driving this increased risk include parental fertilizer, pesticide, solvent, and wood dust exposure. Less consistently reported associations include an increased risk of Ewing sarcoma with parental cigarette smoking and parental occupation in manual labor. Family income does not appear to be associated with the risk of Ewing sarcoma.
Additional attention has focused on features of physical growth and development that might distinguish patients with Ewing sarcoma. Differences in birth weight between patients with Ewing sarcoma and controls have not been observed. Given its peak incidence during adolescence, some groups have investigated differences in the onset of puberty between patients with Ewing sarcoma and controls. One group reported that boys with Ewing sarcoma began shaving earlier than boys without Ewing sarcoma. Other differences related to development of secondary sex characteristics have not been observed. At the time of diagnosis, patients with Ewing sarcoma do not seem to be taller than their peers.
Several viruses have been suggested as playing a role in the pathogenesis of Ewing sarcoma. One group reported that the adenovirus E1A gene could specifically induce the chromosomal translocation characteristic of these tumors. Follow-up studies failed to corroborate this finding. Based on increased Epstein-Barr virus titers in patients with Ewing sarcoma, other groups evaluated Ewing tumors for Epstein-Barr virus infection and found no viral involvement. Both BK and SV40 viral sequences have been variably identified in cases of Ewing sarcoma, but the functional importance of these findings remains unknown.
Ewing sarcoma rarely develops as a second malignancy. Several case reports describe the development of Ewing sarcoma years after the successful treatment of children with a range of hematologic malignancies and solid cancers. More formal studies indicate that Ewing sarcoma as a second malignancy occurs only infrequently. In a population-based study, 2.1% of all Ewing sarcoma cases arose as second malignancies. In a large institutional review, only 1.3% of second malignancies following treatment of pediatric cancer were Ewing sarcoma or PNET. Several large series of patients with secondary bone sarcomas have reported only a small number of patients with Ewing sarcoma. These secondary Ewing tumors do not appear to be radiation-related. In addition, the wide range of primary malignancies described in these patients suggests that these tumors do not arise as part of a specific cancer predisposition syndrome.
In the mid-1980s the presence of a recurrent reciprocal chromosomal translocation, t(11;22)(q24;q12), was reported in Ewing sarcoma tumors and cell lines. The same translocation was also identified in PNET, supporting the hypothesis that these two tumors represented the same disease exhibiting varying levels of differentiation along a neuronal pathway. Subsequent studies demonstrated that t(11;22)(q24;q12) was present in approximately 83% of Ewing sarcomas.
The early analysis of translocations in Ewing sarcoma demonstrated that the derivative chromosome 22 was always maintained as these tumors and cell lines undergo clonal evolution, whereas the derivative chromosome 11 could be lost. This indicated that the “22;11” derivative is the key chromosomal abnormality, whereas the reciprocal “11;22” is unnecessary. The t(11;22)(q24;q12) is a tumor-specific or somatic mutation because it is not found in the germline DNA of patients.
In addition to the t(11;22)(q24;q12) rearrangement (and related translocations with similar molecular consequences, discussed later), other chromosomal abnormalities have also been reported in Ewing sarcoma. A careful analysis of these sometimes complex karyotypes has revealed a small group of recurrent, nonrandom chromosomal abnormalities. Gains (usually trisomies) of chromosome 8 may be observed in up to 50% of cases, and gains of chromosomes 12 and 1q are seen in approximately 25% of cases. Gains in chromosome 20 have also been reported in 10% to 20% of cases. Interestingly, a chromosomal translocation that is unrelated to the t(11;22)(q24;q12) has been observed in approximately 20% of cases: der(16)t(1;16). This translocation is often found as an unbalanced rearrangement and thus results in partial gains (trisomies and occasionally tetrasomies) of 1q and losses (monosomies) of 16q. The breakpoints of this translocation appear to be variable and range from q11 through q32 on chromosome 1 and from q11.1 through q24 on chromosome 16. This suggests that the translocation does not alter particular target genes at the breakpoint but rather introduces gains of 1q and losses of 16q. Finally, losses at 1p36 have also been observed. More recent high-resolution copy number studies have identified copy number alterations in specific genes or regions of the genome, although the role of specific genomic gains or losses has yet to be determined. There has also been a report of genomic instability, as measured by microsatellite instability and loss of heterozygosity, in Ewing sarcoma. This finding, however, is somewhat controversial, insofar as a second group was not able to replicate the microsatellite instability findings.
A major advance in the understanding of the pathogenesis of Ewing sarcoma occurred when the breakpoint of the (11;22)(q24;q12) translocation was cloned in 1992. The translocation breakpoint was localized roughly in the middle of two genes: EWSR1 and FLI1 . EWSR1 had not been previously identified and so was named on the basis of its involvement in the Ewing sarcoma translocation breakpoint ( Ew ing s arcoma r earrangement domain 1 ). FLI1 had been previously identified in mice as the F riend L eukemia Virus i ntegration site. As a result of the translocation event, these two genes become fused and the expressed transcript encodes the EWS/FLI fusion protein ( Fig. 61-3 ).
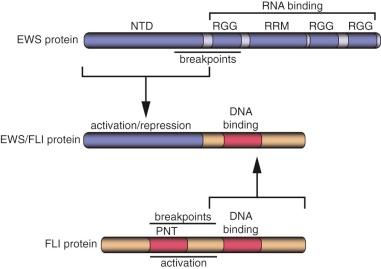
EWSR1 encodes the EWS protein (see Fig. 61-3 ). Wild-type EWS contains 656 amino acids and appears to be ubiquitously expressed. EWS is primarily localized to the nucleus, although it has also been detected in the cytoplasm and on the cell surface. The amino terminus of EWS (sometimes referred to as NTD-EWS) contains 285 amino acids consisting of 31 pseudorepeats rich in tyrosine, glutamine, serine, threonine, glycine, alanine, and proline. This region bears some similarity to the carboxyl-terminal domain (CTD) of RNA polymerase II. This similarity suggested that the region could modulate RNA transcription. The carboxyl terminus of EWS contains an RNA recognition motif (an RRM domain), as well as three arginine-glycine-glycine (RGG) domains that are often found in RNA binding proteins. Indeed, it has been demonstrated, in vitro, that EWS can bind to RNA. These findings suggest that wild-type EWS may be involved in RNA transcription or processing. The carboxyl-terminal region also contains a potential C 2 C 2 zinc finger of uncertain significance. There is evidence that EWS may undergo posttranslational modifications, but the significance of these alterations is not well understood.
EWS is closely related to two additional sarcoma-associated translocation partners, TLS ( t ranslocated in l ipo s arcoma; also called FUS) and TAF15 (also known at TAF II 68, TAF2N, and RBP56), as well as to a Drosophila melanogaster protein named cabeza (also known as SARFH; originally called P19). TLS was first identified in human myxoid liposarcoma in the context of the t(12;16)(q13;p11) translocation, which encodes a TLS/CHOP fusion protein ( Table 61-1 ). TAF15 is a member of the TFIID general transcription complex. It is found fused to NR4A3 (also called CHN, TEC, CSMF, or NOR1) in extraskeletal myxoid chondrosarcomas and to ZNF384 (also called CIZ or NMP4) in acute leukemias (see Table 61-1 ). TAF15 also has significant homology and domain organization to EWS and TLS. The three human proteins are sometimes described as the TET family of proteins ( T LS, E WS, T AF15) or the FET family ( F US, E WS, T AF15). Although the normal function of these proteins is not completely understood, some evidence suggests that they may be involved in RNA splicing, gene expression, microRNA processing, and formation of RNA-containing intracellular granules. Interestingly, constitutional mutations in TLS result in familial amyotrophic lateral sclerosis (ALS), and mutations in TAF15 may also be associated with the disease.
| Tumor Type | Translocation | Fusion Gene |
|---|---|---|
| Ewing sarcoma | t(11;22)(q24;q12) | EWSR1/FLI1 |
| t(21;22)(q22;q12) | EWSR1/ERG | |
| t(7;22)(p22;q12) | EWSR1/ETV1 | |
| t(17;22)(q12;q12) | EWSR1/ETV4 | |
| t(2;22)(q35;q12) | EWSR1/FEV | |
| t(16;21)(p11;q22) | TLS/ERG | |
| t(2;16)(q35;p11) | TLS/FEV | |
| Ewing-like sarcoma | t(20;22)(q13;q12) | EWSR1/NFATC2 |
| (NB: can occur in ring chromosome and may be amplified) | EWSR1/POU5F1 | |
| t(6;22)(p21;q12) | EWSR1/SMARCA5 | |
| t(4;22)(q31;q12) | EWSR1/ZSG | |
| Submicroscopic inv(22) in t(1;22)(p36.1;q12) | ||
| t(2;22)(q31;q12) | EWSR1/SP3 | |
| t(4;19)(q35;q13) | CIC/DUX4 | |
| inv(X)(p11.4;p11.22) | BCOR/CCNB3 | |
| Clear cell sarcoma | t(12;22)(q13;q12) | EWSR1/ATF1 |
| Desmoplastic small round cell tumor | t(11;22)(p13;q12) | EWSR1/WT1 |
| Extraskeletal myxoid chondrosarcoma | t(9;22)(q22;q12) | EWSR1/NR4A3 |
| t(9;17)(q22;q11) | TAF15/NR4A3 | |
| t(9;15)(q22;q21) | TCF12/NR4A3 | |
| Myxoid liposarcoma | t(12;16)(q13;p11) | TLS/DDIT3 |
| t(12;22)(q13;q12) | EWSR1/DDIT3 | |
| Angiomatoid fibrous histiocytoma | t(12;16)(q13;p11) | TLS/ATF1 |
| Low grade fibromyxoid sarcoma | t(7;16)(q33;p11) | TLS/CREB3L2 |
| Acute myelogenous leukemia | t(16;21)(p11;q22) | TLS/ERG |
| Acute myelogenous, lymphoblastic, or undifferentiated leukemia | t(12;22)(p13;q12) | EWSR1/ZNF384 |
| t(12;17)(p13;q11) | TAF15/ZNF384 |
* Gene names are provided rather than their corresponding protein names.
FLI is a member of the ETS family of transcription factors. This family is defined by the presence of an ETS type DNA-binding domain and consists of 27 different members in humans. As with most transcription factors, ETS family members have a modular architecture with separate domains that contribute DNA binding and transcriptional functions. The DNA-binding ETS domain of FLI is present in the carboxyl-terminal portion of the protein (see Fig. 61-3 ). Most ETS family members (including FLI) bind to sequences containing a GGAA or GGAT core sequence. In addition to DNA-binding function, FLI contains two separate transcriptional activation domains, one in the amino terminus and a second in the carboxyl terminus (distal to the ETS DNA-binding domain; see Fig. 61-3 ). Wild-type FLI also contains a PNT (pointed) domain in the amino terminal portion of the protein. PNT domains are thought to mediate protein-protein interactions.
The normal role of FLI appears to be primarily in the hematopoietic lineage, where it regulates megakaryocytic development. Knockout of murine Fli-1 results in abnormal megakaryocytic differentiation and a loss in vascular integrity that results in embryonic lethality from central nervous system hemorrhage. Hemizygous deletion of FLI is found in patients with Paris-Trousseau or Jacobsen thrombocytopenia syndromes. FLI also appears to be important for vascular development in the mouse and zebrafish. FLI is also expressed in neural crest–derived mesenchyme in both the mouse and quail. This finding is particularly interesting in light of the neural crest phenotype of Ewing sarcoma.
FLI itself can function as an oncogene, at least in the hematopoietic context. Indeed, the murine Fli locus was first defined as a predominant F riend mu rine l eukemia v irus (F-MuLV) insertion site. Thus insertion of F-MuLV at the Fli locus results in upregulation of the Fli gene and subsequent development of erythroleukemia. The oncogenic activity of FLI appears to be limited to the hematopoietic system as introduction of the protein into other models of oncogenesis, such as the NIH3T3 cell model, does not result in the transformed phenotype.
EWS/FLI consists of the amino terminus of EWS fused, in frame, to the carboxyl terminus of FLI (see Fig. 61-3 ). The EWS portion retained in the fusion contains the pseudorepeat domain that harbors a strong transcriptional activation function as well as a transcriptional repression function. The RNA-binding domain of EWS is lost in the fusion protein and instead is replaced by the portion of FLI that contains its ETS DNA-binding domain.
EWS/FLI functions as an oncoprotein. EWS/FLI can induce NIH3T3 cells (an immortalized mouse fibroblast cell line) to exhibit both anchorage independent growth and growth as tumors when injected subcutaneously into immunodeficient mice. Conversely, blockade of EWS/FLI expression or function (via antisense RNA, dominant-negative blocking alleles, or RNA interference) causes Ewing sarcoma cells to lose their transformed phenotype. These data demonstrate that EWS/FLI expression is required for the oncogenic phenotype of Ewing sarcoma. Interestingly, it has been reported that after therapy-induced neural differentiation of Ewing sarcoma, EWS/FLI expression may also be lost. Although these data support an important role for EWS/FLI expression in Ewing sarcoma proliferation and transformation, they do not address whether EWS/FLI formation is the first step in Ewing sarcoma development or whether other oncogenic events occur first and are followed by EWS/FLI formation.
There are three main consequences of the t(11;22)(q24;q12) rearrangement. First, it places the transcriptional regulation of the EWS/FLI fusion under the control of the EWSR1 promoter. This is important because the activity of the FLI1 promoter is limited to hematopoietic (and perhaps neural crest) lineages, whereas the EWSR1 promoter appears to be active in most, if not all, cell types (see previous sections). Second, the translocation results in a loss of one wild-type EWSR1 allele and one wild-type FLI1 allele. Loss of one FLI1 allele is probably of no consequence because wild-type FLI1 is not expressed in Ewing sarcoma. Whether loss of one EWSR1 allele is important in Ewing sarcoma development is not known. The third consequence of the translocation is that it creates the EWS/FLI fusion protein. This is important because although EWS/FLI functions as an oncoprotein, neither wild-type EWS nor wild-type FLI could induce transformation of NIH3T3 cells, thus demonstrating a gain of function for the EWS/FLI fusion. Structure-function analyses of EWS/FLI demonstrated that both the amino terminal EWS domain and the ETS DNA-binding domain of FLI were required for oncogenic transformation of NIH3T3 cells and Ewing sarcoma cells. These results indicate that the fusion protein functions as an aberrant transcription factor. Comparisons of EWS/FLI with wild-type FLI demonstrated that the EWS domain included in the fusion protein could function as a much stronger transcriptional activation domain than the domain of FLI that was lost in the fusion. These results suggested that EWS/FLI induced oncogenic transformation by binding to target genes through its ETS DNA-binding domain and upregulating the expression of those genes through its EWS domain.
The mechanisms by which the EWS portion of the fusion functions as a transcriptional activation domain are only now beginning to be understood. Because wild-type EWS has been shown to interact with TFIID, RNA polymerase II, or the p300/CBP coactivators, it appears likely that these interactions are also important for EWS/FLI function. RNA helicase A (RHA) has also been shown to be important for the transcriptional activity of EWS/FLI. Thus RHA may also function as a coactivator for gene expression mediated by the fusion protein.
More recently, it has been appreciated that EWS/FLI also functions as a transcriptional repressor. For example, analysis of the transcriptional program triggered by EWS/FLI in Ewing sarcoma cells has revealed that more genes are downregulated by the fusion than are upregulated. Additional analysis has demonstrated that EWS/FLI functions as a direct transcriptional repressor at some target genes, like LOX and TGFBR2 . The mechanism of repression appears to be through binding of a transcriptional corepressor complex called NuRD, that includes nucleosome remodeling function, as well as histone deacetylase and histone lysine-specific demethylase activities. Both transcriptional activation and repression functions are required for the oncogenic activity of EWS/FLI in Ewing sarcoma cells.
Analysis of EWS/FLI binding sites in the human genome revealed that the fusion protein binds to sites that contain “classic” ETS-type high-affinity sequences (e.g., ACCGGAAGTG). However, a more interesting and unusual finding was that EWS/FLI was also found to bind to GGAA-containing microsatellite sequences. EWS/FLI binds to these microsatellites and uses them as response elements to activate the expression of target genes, and the transcriptional function is proportional to the length of the microsatellite. This raises the intriguing possibility that polymorphisms in GGAA-microsatellites associated with key EWS/FLI target genes (e.g., NR0B1, CAV1, or GSTM4 ) may alter susceptibility to Ewing sarcoma or perhaps outcome in patients with the disease. It has also been suggested that the lack of GGAA-microsatellites near critical EWS/FLI target genes, such as NR0B1 , in organisms other than humans might be the reason that only humans develop Ewing sarcoma.
In addition to EWS/FLI, other less frequent Ewing sarcoma translocation breakpoints were cloned and revealed a very similar structural organization (see Table 61-1 ). As described, approximately 83% of Ewing sarcoma tumors contain an (11;22)(q24;q12) translocation. Approximately 10% of cases demonstrate an alternate translocation, t(21;22)(q22;q12). Cloning of the breakpoint of this rearrangement revealed that EWS was fused, in frame, to another member of the ETS family, ERG. As in the case of EWS/FLI, the EWS/ERG fusion contains the amino terminus of EWS fused to the carboxyl terminus of ERG, which harbors its ETS DNA-binding domain. Interestingly, the ETS DNA-binding domains of FLI and ERG are 98% identical at the amino acid level. Subsequent investigations demonstrated that EWS/ERG also functioned as an oncoprotein in the NIH3T3 cell model. In subsequent years additional translocations were identified in Ewing sarcoma, including a t(7;22)(p22;q12), t(17;22)(q12;q12), and t(2;22)(q33;q12). Cloning of transcripts from these translocations demonstrated that in each case EWS was fused to another member of the ETS family. Currently, five EWS/ETS fusions have been described (see Table 61-1 ). It is assumed that all EWS/ETS fusion proteins bind similar (if not identical) target genes to induce oncogenic transformation.
Overall, most cases of Ewing sarcoma appear to harbor fusions between EWS and various ETS family members. Rarely, fusions between TLS and ETS family members may be present in Ewing sarcoma instead, as shown by the identification of TLS/ERG fusions in four patients with Ewing sarcoma and the identification of a TLS/FEV fusion in another case (see Table 61-1 ). These findings highlight the similarities across the TET family and across the ETS family. Perhaps the most generic manner of describing the fusion proteins in Ewing sarcoma would be as “TET/ETS fusions.” Because EWS/FLI is the most common fusion identified in Ewing sarcoma, most of the experimental work has been performed using this fusion protein. Experimental findings based on EWS/FLI have only occasionally been confirmed using other Ewing sarcoma fusion proteins.
In addition to variability of fusion partners, variability also exists in the exonic structure of some of the fusion proteins ( Fig. 61-4 ). The breakpoints found in Ewing sarcoma translocations occur in the introns of EWSR1 and FLI1 (and presumably the other ETS factors as well, although in most cases this has not been experimentally confirmed). The breakpoints in the EWSR1 gene are found in one of two regions (3 kb and 1.2 kb in size) present in an approximately 6 kilobase (kb) region that includes four of the gene's 16 introns. The breakpoints in FLI1 occur over an approximately 50-kb region, which includes six of eight FLI introns. Splicing events join adjacent exons together in the fusion transcript. As a result of this variability in breakpoint and subsequent RNA splicing, at least 10 different EWS/FLI isoforms are generated (see Fig. 61-4 ).
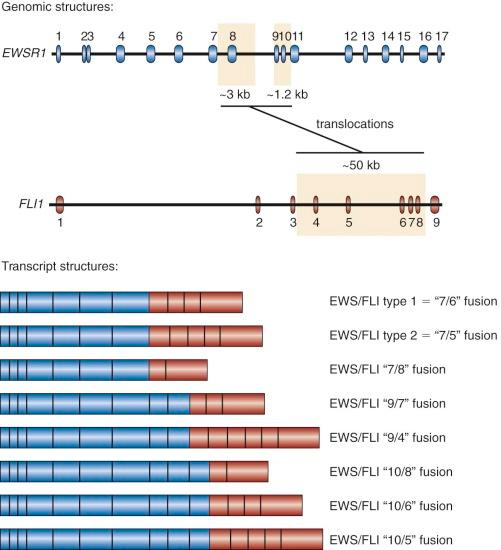
The most common Ewing sarcoma translocation fuses exon 7 of EWSR1 to exon 6 of FLI1 . This “7/6” fusion is commonly referred to as a “type 1” EWS/FLI fusion and accounts for approximately 50% to 60% of EWS/FLI fusions. A “type 2” fusion joins exon 7 of EWSR1 to exon 5 of FLI1 and is the second most common EWS/FLI fusion, accounting for approximately 20% to 30% of EWS/FLI fusions. Laboratory studies suggested that the type 1 EWS/FLI fusion was a weaker oncoprotein than the type 2 fusion, and originally it was thought to have prognostic significance. However, these functional differences were not reflected on changes in gene expression between type 1 and type 2 fusions, and large-scale clinical correlative data did not indicate a prognostic difference based on fusion subtype.
Although there is general agreement regarding the designation of the type 1 and type 2 fusions, there is no generalized nomenclature for the remaining fusions, and so these are best described in terms of which exons are fused to one another. In addition to variability in the EWS/FLI breakpoint, EWS/ERG has also been reported to demonstrate variability based on fusion points, with at least four fusions reported. Because the other fusions are so rare, there is only minimal information as to whether they also show similar levels of breakpoint heterogeneity.
The cell of origin of Ewing sarcoma is unknown, and the subject has generated intense debate. The original description by James Ewing in 1921 suggested that this “round cell sarcoma” was of endothelial origin and hence properly referred to as a “diffuse endothelioma of bone.” Dr. Ewing later suggested that it may arise from perivascular lymphatic endothelium. A hematologic origin was proposed in the early 1970s following an in-depth evaluation of light and electron microscopic characteristics and cytochemistry of both primary tumor and cell culture specimens. In the early 1980s a fibroblastic-mesenchymal derivation was proposed based on, among other things, the patterns of collagen expression of the tumor. In the late 1980s, however, a neural crest derivation was proposed because of the occasional identification of certain neural features, such as the presence of Homer Wright rosettes, neural processes, neurosecretory granules, and neural immunohistochemical markers. Additionally, undifferentiated Ewing sarcoma cell lines can occasionally be induced to express markers of neural differentiation, including the expression of neuritelike elongated processes, neuron-specific enolase (NSE), cholinesterase, and neural filament triplet protein. Interestingly, there are a few examples of neural differentiation of Ewing sarcoma occurring after therapy, which further supports this theory. More recently, it was shown that introduction of EWS/FLI into murine mesenchymal stem cells could result in oncogenic transformation, suggesting that these may be the cell of origin (although the same does not hold true for human mesenchymal stem cells). This hypothesis was further supported by microarray and cell biological studies performed on Ewing sarcoma cell lines in which EWS/FLI expression was reduced using RNA interference techniques. In the absence of EWS/FLI expression, these cells had a gene expression pattern that was reminiscent of mesenchymal stem cells and a cellular structural organization and adhesive and migratory properties that were also consistent with a mesenchymal phenotype. Furthermore, Ewing sarcoma cells with reduced EWS/FLI expression could be induced to differentiate into other mesenchymal cell types, such as fat and bone. This is a characteristic that is shared by mesenchymal stem cells.
It is important to note, however, that the phenotype of Ewing sarcoma (at least the neural crest component) may be a consequence of the (11;22) translocation rather than being related to the tumor's cell of origin. Introduction of EWS/FLI into NIH3T3 murine fibroblasts induces a phenotype that is highly similar to Ewing sarcoma, including the small round blue cell morphology and features of neural differentiation. Similarly, expression of EWS/FLI in human rhabdomyosarcoma cells, neuroblastoma cells, or even normal human fibroblasts causes those cells to express genes that are similar to the genes expressed in Ewing sarcoma. This suggests that EWS/FLI may cause transdifferentiation of its target cells. Thus the neural crest phenotype of Ewing sarcoma may be induced by EWS/FLI.
Transcriptional profiling experiments (e.g., those using oligonucleotide or complementary DNA microarrays and, more recently, RNA sequencing) in Ewing sarcoma tumors and cell lines have allowed for a comprehensive analysis of the gene expression patterns associated with this disease. One of the earliest studies compared Ewing sarcoma tumors and cell lines with other small round cell tumors of childhood, including neuroblastoma, non-Hodgkin lymphoma, and rhabdomyosarcoma. These studies demonstrated that these histologically similar tumors could be distinguished from one another using gene expression data. A more recent analysis compared the expression of approximately 180 sarcoma tumor samples and again showed that tumors could be distinguished using gene expression patterns. Additional studies in which the gene expression pattern of Ewing sarcoma was compared with that of a variety of normal tissues supported the neural crest derivation but also suggested some similarity to endothelial cells. As discussed in the preceding section, analysis of some model systems has suggested a mesenchymal stem cell origin. A definitive understanding of the cell of origin of these tumors will require additional studies. It should be noted that as of yet no genetically engineered mouse model of Ewing sarcoma has been reported, and therefore tissue-specific oncogene expression or lineage-tracing approaches have not been used to identify the Ewing sarcoma cell of origin.
Because most studies have suggested that EWS/FLI functions as a transcription factor, the two biggest questions in the field have been (1) what genes are dysregulated by EWS/FLI and (2) what are the roles of these gene targets in oncogenic transformation? These questions are beginning to be answered. Because the cell of origin of Ewing sarcoma is unknown, most laboratory-based studies of EWS/FLI have used heterologous cell types with varying results. More recent approaches have benefited from the use of “knockdown” (reduction) of gene expression using RNA interference (RNAi). The RNAi technique has allowed for the analysis of EWS/FLI in patient-derived Ewing sarcoma cell lines themselves, thus avoiding the concerns associated with using heterologous cell types.
Gene expression analysis of Ewing sarcoma cells in which EWS/FLI has been knocked down has allowed a comprehensive identification of the thousands of genes that are modulated by the fusion protein. For example, increased expression of NKX2.2 and NR0B1 are required for the oncogenic phenotype of Ewing sarcoma, although the specific mechanisms by which these transcriptional regulators facilitate the transformed phenotype remains poorly understood. Similarly, repression of TGFBR2 (the receptor for transforming growth factor β) and lysyl oxidase (LOX) by EWS/FLI are also required for tumorigenicity of Ewing sarcoma cells.
Gene expression studies also demonstrated that EWS/FLI represses the expression of the insulin-like growth factor binding protein 3 (IGFBP-3). IGFBP-3 appears to be involved in Ewing sarcoma apoptosis, and thus decreased expression of this protein appears to prevent apoptosis. These data complement a growing body of literature that supports a critical role for IGF-1 signaling in Ewing sarcoma development. For example, it was shown that Ewing sarcoma tumors and cell lines express IGF-1 and IGF receptors. IGF-1 and its receptor appear to function in an autocrine/paracrine fashion in Ewing sarcoma, and disruption of this signaling pathway blocks various functions that contribute to tumorigenesis. This occurs both in Ewing sarcoma cell lines as well as in model systems. Indeed, disruption of this pathway has shown some efficacy as a molecularly targeted therapeutic approach for this disease (discussed later).
NPY1R was identified as an EWS/FLI target gene through microarray analysis. Neuropeptide Y (NPY) is a small polypeptide neurotransmitter with diverse functions. Engagement of NPY receptors by NPY in Ewing sarcoma cell lines is growth inhibitory, suggesting a potential therapeutic role for these receptors. This result was surprising, insofar as Ewing sarcoma cells also express the NPY ligand. It appears likely that Ewing sarcomas are protected from the growth-inhibitory effect of NPY/NPY1R interactions by the expression of dipeptidyl peptidase IV, which cleaves NPY into a form that is unable to bind NPY1R. This cleaved form, NPY 3-36 , functions as an angiogenic factor by binding to NPY2R receptors on endothelial cells, thus providing a likely explanation for the maintenance of this growth-inhibitory autocrine pathway in Ewing sarcoma cells.
Another protein that has been shown to be upregulated by EWS/FLI is vascular endothelial growth factor (VEGF). VEGF levels are increased in Ewing sarcoma cell lines and xenografts. VEGF levels are also increased in the blood of patients with Ewing sarcoma and VEGF has been shown to be expressed in 55% of Ewing sarcoma tumor samples by immunohistochemistry. In the case of Ewing sarcoma, VEGF may induce not only angiogenesis but also vasculogenesis. Blockade of VEGF function in Ewing sarcoma model systems prevents tumor growth. These preclinical studies demonstrate the potential for VEGF inhibition as a therapeutic strategy for Ewing sarcoma.
Most, but not all, studies of Ewing sarcoma have shown that these tumors typically express telomerase activity. Interestingly, it appears that hTERT, the catalytic subunit of the telomerase holoenzyme, is upregulated by EWS/ETS proteins. This upregulation appears to be caused by specific binding of the ETS portion of the fusion to the TERT promoter. These data suggest that EWS/ETS fusions regulate telomerase activity by direct activation of the TERT promoter.
One key protein whose expression may be regulated by EWS/FLI is CD99 (also called MIC2). In human fibroblasts engineered to express EWS/FLI, the expression of CD99 closely mimicked that of the fusion protein. In the early 1990s it was found that CD99 is expressed at high levels in Ewing sarcoma cells but is uncommonly found on other tumor types. The CD99 antigen is recognized by a variety of monoclonal antibodies, including 12E7, HBA71, and O13, and these antibodies have an important role in the diagnosis of Ewing sarcoma (discussed later). The CD99 antigen is a membrane glycoprotein that plays a role in T-cell development and activation and migration of monocytes through endothelial junctions. In Ewing sarcoma CD99 inhibits neural differentiation and thus contributes to oncogenic transformation. Significantly, emerging data suggest that antibody-mediated engagement of CD99 may eventually be exploited as a new therapeutic approach to Ewing sarcoma. CD99 engagement in Ewing sarcoma results in apoptosis of the tumor cells. This effect enhances tumor cell killing by chemotherapeutic agents. Gene expression studies of CD99-mediated apoptosis suggested a role for the cytoskeletal protein zyxin in this process, and this finding was confirmed through functional studies. The identification of zyxin in this process was interesting in light of subsequent studies demonstrating that zyxin acts as a tumor suppressor in Ewing sarcoma. Taken together, these studies suggest that CD99 may be an effective therapeutic target in Ewing sarcoma by modulating cytoskeletal actin function.
Multiple potentially cooperating mutations in addition to TET/ETS fusions have been identified in Ewing sarcoma. The most widely assessed are mutations in the p53 and RB pathways. The p53 pathway includes proteins such as p14 ARF and MDM2, as well as p53 itself. When expressed in some heterologous systems, such as primary human fibroblasts, EWS/FLI induces the expression of p53. This finding suggests that there is selective pressure to inhibit the p53 pathway. This hypothesis appears to be at least partially true. Deletions in the CDKN2A locus (commonly called the INK4a locus encoding both the p14 ARF and p16 INK4a proteins; note that p14 ARF is involved in the p53 pathway, whereas p16 INK4a is involved in the RB pathway) have been identified in approximately 10% to 30% of primary Ewing sarcoma tumors and more than 50% of Ewing sarcoma cell lines. Amplifications or overexpression of MDM2 is occasionally seen in Ewing sarcoma. Mutations in p53 are found in 5% to 20% of Ewing sarcomas. Taken together, mutations in the p53 pathway may be present in nearly 70% of Ewing sarcomas, and these mutations may have prognostic significance in this disease (discussed later). Because there are likely additional components of the pathway yet to be identified or analyzed, the frequency of alterations in this pathway may be even higher.
The RB pathway regulates the cell cycle and includes proteins such as CDK4, D-type cyclins, and p16 INK4a . Mutations have been found in RB itself, and deletions of the INK4a locus have been documented as well in Ewing sarcoma. As was observed in the case of the p53 pathway, mutations in the RB pathway also appear to be relatively common in this disease.
Whereas mutations in the p53 and RB pathways are relatively common in Ewing sarcoma, mutations in other genes or pathways have only rarely been identified. For example, mutations in BRAF (which is also a member of the RAS/MAPK pathway) have been identified in 1 of 22 cell lines examined. No mutations in other components of the pathway, including RAS or receptor tyrosine kinases, have been identified, as of yet. This suggests that the RAS/MAPK pathway is only rarely activated by mutation in this disease.
Although the RAS/MAPK pathway does not appear to be frequently activated by mutation, there is some evidence for a requirement of pathway activation in Ewing sarcoma tumor development. In the NIH3T3 model system, it was shown that expression of EWS/FLI induced phosphorylation of the ERK1 and ERK2 proteins (downstream members of the MAPK pathway). This effect may result from autocrine stimulation of these transformed cells by PDGF-C, which was also shown to be induced by EWS/FLI expression in the NIH3T3 model, and expressed in Ewing sarcoma cell lines and tumor specimens. There is some controversy as to whether PDGF-C plays a role in the human disease, as PDGFR-α, which is required for PDGF-C responsiveness, is not expressed in Ewing sarcoma. Although PDGF-C may not be involved in Ewing sarcoma, inhibition of the MAPK pathway does block oncogenic transformation mediated by EWS/FLI in the NIH3T3 model. This finding was not replicated in Ewing sarcoma cells, thus raising the question as to whether this is an NIH3T3-specific pathway.
Many members of the Wnt signaling pathway are expressed in Ewing sarcoma, including soluble Wnt ligands, Wnt receptors (frizzled), and Wnt coreceptors (LRP5/6). Additionally, microarray analysis of EWS/FLI target genes has demonstrated downregulation of the Wnt inhibitor DKK1 and upregulation of various Wnt signaling components. These data suggest that Wnt signaling occurs in Ewing sarcoma. In contrast, analysis of the pathway in Ewing sarcoma cell lines in tissue culture has not demonstrated evidence of active Wnt autocrine signaling. If the pathway is involved in Ewing sarcoma, it seems likely that it is important for processes such as migration and metastasis, rather than proliferation and transformation. Current studies have not addressed this possibility.
Finally, genes involved in chromosomal stability or chromatin remodeling have also been implicated in Ewing sarcoma. For example, the STAG2 protein helps regulate chromosomal separation during mitosis and was found to be deleted in a number of human tumor types, including Ewing sarcoma. It was suggested that this may be a mechanism of chromosomal instability in Ewing sarcoma and other tumors. Deletion of SMARCB1 (encoding the SMARCB1/INI1/SNF5 protein) was found in approximately 10% of Ewing sarcoma cases. SMARCB1 is a member of the SWI/SNF chromosome remodeling complex whose deletion is tightly associated with atypical teratoid/rhabdoid tumor. Whether SMARCB1-deleted Ewing sarcoma tumors represent a unique subset of Ewing sarcoma or instead indicate that alterations in chromatin remodeling complexes are widespread in Ewing sarcoma is currently unknown. Interestingly, a newly identified chromosomal rearrangement, t(4;22)(q31;q12), encodes an EWS/SMARCA5 fusion protein in tumors that appear consistent with Ewing sarcoma, supporting the idea that alterations in this chromatin remodeling complex may indeed be widespread in this disease. Another possibility is that the EWS/SMARCA5 fusions are found in tumors that are similar, but not identical, to Ewing sarcoma. Indeed, the most recent World Health Organization guidelines have named these (and other non-TET/ETS translocation-bearing tumors that have similar appearance to Ewing sarcoma; see Table 61-1 ) as Ewing-like sarcomas.
As described previously, mutations in the p53 pathway are relatively common in Ewing sarcoma. These mutations likely confer resistance to the growth effects of TET/ETS fusions in the disease and may also protect against apoptosis. Whereas p53 is an important mediator of apoptosis, other signaling pathways are involved as well. These other pathways have been evaluated in Ewing sarcoma in a number of studies.
Both Fas and the Fas ligand (FasL) are expressed in the majority of Ewing sarcomas. The status of the FasL in Ewing sarcoma is somewhat controversial; one study reported soluble FasL in Ewing sarcoma–conditioned media, whereas another study reported that the FasL is only found intracellularly and thus is not accessible to cell surface receptor. In terms of the receptor, Fas at the cell surface can be functional, but Ewing sarcoma cells can be divided into Fas-sensitive, Fas-inducible, and Fas-resistant groups. Fas-sensitive lines are killed by coincubation with a FasL-expressing effector cell. Fas-inducible lines are killed by this effector cell line only after pretreatment with interferon-γ (IFN-γ) or cycloheximide (or both). The Fas-resistant lines were resistant regardless of pretreatment. The differences between sensitive and resistant cells are not completely known but may involve differences in Fas expression or expression of proapoptotic or antiapoptotic mediators, such as BAD and BAR. Strategies to improve Fas-mediated killing of Ewing sarcoma cells have been investigated, such as allowing for accumulation of more FasL through the use of metalloproteinase inhibitors and upregulation of Fas by administration of interleukin-12.
In addition to Fas/FasL, evaluations of tumor necrosis factor–related apoptosis-inducing ligand and tumor necrosis factor α as inducers of Ewing sarcoma apoptosis have also been conducted, in vitro and in vivo, alone and in combination with other agents. Taken together, these data support a potential use of proapoptotic ligands in the treatment of Ewing sarcoma. However, the variability of the effects seen demonstrate the need for an ongoing mechanistic evaluation of these pathways in this disease.
The identification of t(11;22)(q24;q12) and subsequent cloning of the EWS/FLI fusion protein represented a major advance in the understanding of the pathogenesis of Ewing sarcoma. EWS/FLI appears to function primarily as an aberrant transcription factor to dysregulate gene targets involved in the development of this disease. Multiple gene targets and cooperating molecular pathways have been identified. These represent a wide array of functions, including growth factor signaling, survival and apoptosis pathways, angiogenesis, cellular immortalization, and cellular differentiation. Despite these myriad molecular functions, a unified mechanistic understanding of Ewing sarcoma development has not yet been reached. Similarly, the cell of origin of the disease has not yet been clearly identified. Nonetheless, there is great hope that a detailed understanding of the molecular mechanisms involved in Ewing sarcoma development will lead to new diagnostic, prognostic, and therapeutic approaches for this disease.
The most common presenting symptoms of Ewing sarcoma are pain, a palpable mass, or both. In one series 89% of patients with Ewing sarcoma reported pain at the time of initial presentation. Pain does not necessarily occur at night; this pattern was reported in only 19% of patients in one series. Patients may report that their pain began at the same time as a minor musculoskeletal injury. This presentation, reported in approximately 25% of patients, may delay the diagnosis. Unlike patients with osteosarcoma, patients with Ewing sarcoma may report constitutional symptoms such as fever and weight loss. Approximately 15% to 20% of patients have fever at presentation. Other symptoms depend on the site of disease. For example, 40% to 94% of patients with paraspinal tumors have presenting symptoms of spinal cord compression. Patients with large pelvic tumors may complain of an alteration in voiding. Extensive bone marrow metastatic disease may cause symptoms related to anemia, thrombocytopenia, or neutropenia. Patients may have symptoms for many weeks or months before seeing a physician for evaluation, with most studies reporting an average time from symptom onset to diagnosis of 3 to 5 months.
Ewing sarcoma can arise from any bone but has a predilection for pelvic bones and long bones of the leg. Approximately 45% of Ewing sarcomas develop in the axial skeleton. More than half of these axial tumors arise from pelvic bones, and a quarter of axial tumors arise from a rib. Up to 30% of Ewing sarcomas arise from long bones in the leg, and up to 15% of tumors develop in long bones in the arm. Tumors of the hands, feet, and head develop only rarely, accounting for 10% or fewer of all tumors. The distribution of primary tumor site does not vary among racial groups. Soft tissue tumors may arise in any location, although axial sites predominate.
Laboratory studies in patients with newly diagnosed Ewing sarcoma may reveal nonspecific abnormalities. More than a third of patients have an elevated erythrocyte sedimentation rate (ESR) at the time of diagnosis. Similarly, serum lactate dehydrogenase (LDH) levels are elevated in approximately one third of patients. Laboratory manifestations of tumor lysis syndrome are uncommon with these tumors. Unlike osteosarcoma, alkaline phosphatase is not typically elevated in patients with Ewing sarcoma of the bone. Patients with advanced bone marrow metastatic disease may have anemia, thrombocytopenia, and neutropenia detected on a complete blood count. In one large series 12% of patients had anemia even in the absence of metastatic disease.
Box 61-1 summarizes the differential diagnosis of Ewing sarcoma. In most cases of Ewing sarcoma of the bone, the main alternative diagnosis is osteosarcoma. Table 61-2 displays features that may help differentiate Ewing sarcoma of the bone from osteosarcoma. Compared with osteosarcoma, Ewing sarcoma is more likely to occur in the axial skeleton. Those Ewing tumors that arise in long bones have a greater tendency to occur in the diaphysis, compared with the tendency of osteosarcoma to arise in the metaphysis of long bones. Patients with Ewing sarcoma are more likely to report constitutional symptoms than are patients with osteosarcoma. Other malignant tumors in the differential diagnosis of Ewing sarcoma of the bone include primary bone lymphoma, giant cell tumor of the bone, and bone metastasis of an extraosseous malignancy. The differential diagnosis of Ewing sarcoma arising from the soft tissues is more broad because the site of origin for these tumors is not restricted to the bone. The site of origin may help narrow the differential diagnosis. Paraspinal tumors may be confused with neuroblastoma, particularly in younger patients. Pelvic tumors may suggest rhabdomyosarcoma or malignant germ cell tumor. Lymphoma and other soft tissue sarcomas can arise at any site and should be included in the differential diagnosis.
Osteosarcoma
Primary bone lymphoma
Ewing-like sarcoma
Langerhans cell histiocytosis
Osteomyelitis
Metastasis of an extraosseous malignancy
Benign bone tumor
Osteoblastoma
Chondrosarcoma
Giant cell tumor of the bone
Rhabdomyosarcoma
Nonrhabdomyosarcoma soft tissue sarcoma
Lymphoma
Benign soft tissue tumor
Ewing-like sarcoma
Neuroblastoma
Malignant germ cell tumor
| Ewing Sarcoma | Osteosarcoma | |
|---|---|---|
| Age distribution | Peak in adolescence Occurs in young children Very rare in adults >40 years |
Peak in adolescence Very rare in children <5 years Occurs in older adults |
| Racial distribution | Rare in patients of African or East Asian ancestry | No racial predilection |
| Predisposing factors | Not associated with radiation No known familial predisposition |
Radiation exposure Li-Fraumeni syndrome History of retinoblastoma |
| Constitutional symptoms | Yes | No |
| Involved bones | Both flat and long bones | Long bones more common |
| Location in long bones | Diaphyseal more common | Epiphyseal/metaphyseal more common |
| Type of periosteal reaction | Laminated/layered “Onion-skinning” | Spiculated “sunburst” |
| Laboratory findings | Normal alkaline phosphatase Abnormal CBC if marrow disease |
Elevated alkaline phosphatase Normal CBC |
| Histologic findings | Small round blue cells No malignant osteoid |
Malignant spindle cells Malignant osteoid |
Reports from several cooperative group studies have defined the metastatic behavior of Ewing sarcoma. Approximately 25% of patients exhibit distant metastases at presentation. Rates of metastatic disease do not appear to differ between patients with skeletal tumors and those with soft tissue primary tumors. Patients with pelvic primary tumors have an increased incidence of metastatic disease at initial diagnosis. The lung is the most common metastatic site, with pulmonary dissemination reported in 50% to 60% of patients with metastatic disease. Patients with isolated pulmonary metastasis represent between 26% and 36% of cases of metastatic disease. Patients usually have multiple lung nodules identified in both lungs.
The bone is the next most common metastatic site at presentation. One group reported cortical bone metastases in 43% of patients with metastatic disease. A large cooperative group reported that the bone marrow is involved in approximately 19% of metastatic patients at initial diagnosis. A smaller series noted bone marrow involvement in 52% of patients with metastatic disease. This group performed up to 10 bone marrow aspirates on patients at initial presentation, suggesting that routine sampling may underestimate the incidence of bone marrow involvement. Brain metastasis at initial diagnosis appears to be extremely uncommon. In two series of patients with brain metastasis from Ewing sarcoma, all cases were reported at the time of disease recurrence and not at initial presentation.
Fewer than 10% of patients have lymph node involvement at initial diagnosis. The incidence of node involvement is higher in patients with soft tissue rather than bone primary tumors. In one population-based study, the incidence of regional node involvement among patients with soft tissue tumors was 12.4% compared with 3.2% in patients with bone primary tumors. The incidence of regional node involvement is also higher among patients with distant metastatic disease.
This metastatic pattern helps determine the appropriate staging evaluation for patients with Ewing sarcoma ( Box 61-2 ). In addition to computed tomography (CT) or magnetic resonance imaging (MRI) scans of the primary tumor, all newly diagnosed patients should be evaluated with a chest CT scan. Assessment for bone metastasis has conventionally been with radiolabeled-technetium bone scan. More recent data indicate that positron emission tomography (PET) scans are superior for this indication (discussed later), prompting wider adoption of PET imaging at initial diagnosis. At least bilateral bone marrow aspirates and biopsies should routinely be performed in newly diagnosed patients, with biopsy more sensitive for detection of marrow involvement compared with aspirate. Attention to regional nodes on imaging studies is particularly warranted in patients with soft tissue primary tumors. These evaluations will indicate whether a patient has metastatic or nonmetastatic Ewing sarcoma at initial diagnosis. This distinction serves as the main staging classification in practice for these patients. Although a staging classification for musculoskeletal tumors has been devised by Enneking, this system is not routinely used in the clinical care of patients with Ewing sarcoma.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here