Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The change in image format from analog to digital has created a revolution. In fields such as photography and radiology, the transfer of imaging technology from film-based to digital formats has seen the bankruptcies of large companies unable to successfully adapt (e.g., Eastman Kodak), and has seen the adoption of digital-based clinical methods to expand capabilities and hence opportunities (e.g., radiology NightHawk services). The digital “revolution” is now upon pathology and early adopters are finding many parallels with radiology, but also new pathology-unique applications and complexities. This chapter will detail the basics of digital imaging, its current and potential uses and the data in support of these applications, and will also discuss the challenges associated with the adoption of digital technology in the cytopathology laboratory.
The adoption of digital methods in pathology has only been possible with recent rapid increases in computer processing power and storage capability, internet and data transfer speed, and new hardware and software development. There are a range of digital applications, from relatively simple, such as picture taking and archiving; to mid-level tasks, such as interactive continuing education, teleconsultation, and quality assurance exercises; and finally to advanced tasks such as image data-mining and the development of “smart” image analysis tools (artificial intelligence) capable of independent decision-making tasks.
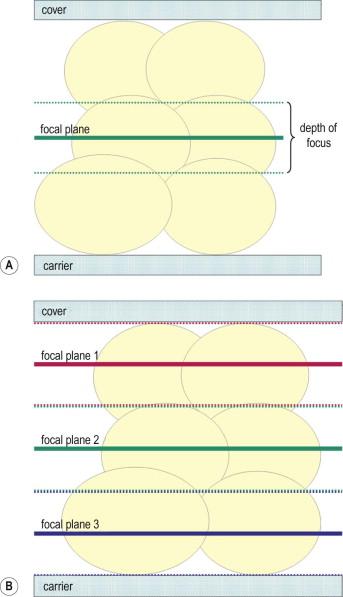
The practice of cytopathology will be involved in the transformation to digital technology; however, compared with digital pathology applications in surgical pathology, the cytologic specimen has unique features that require special equipment functionality and software approaches. The advantage of analog viewing of a specimen is that the entire specimen is present and can be visualized. Digital renditions of analog specimens are essentially “copies” of the specimen. As such, a complete recapitulation of the specimen would require that all aspects of the original specimen are captured in digital format. Complete capture and hence complete recapitulation could occur, but would require capturing depth of field, as well as color, brightness, contrast, and other optical parameters – in their entirety. Histopathology specimens are highly adaptable to simple, single plane digital scanning because of the very nature of their preparation – that being as relatively flat, single plane sections. Thus, single plane high-resolution digital scans recapitulate the original specimen, not completely, but very closely. Cytology specimens, on the other hand, are intrinsically 3-dimensional, requiring special manipulations in order to produce digital renditions that truly recapitulate the original specimen ( Fig. 34-1 ). Therefore, although digital cytology has similar applicability to all the functionality noted above, in practice its adaptability is more challenging.
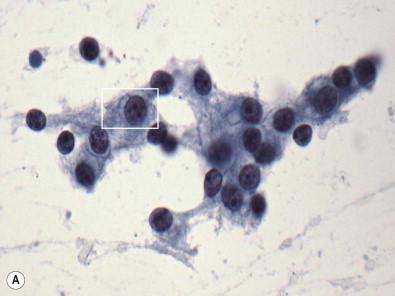
Digital pathology represents a fundamental change in the way in which practitioners view cases. Instead of viewing analog images caused by light waves passing through an actual biologic specimen via an optics-based device (i.e., a microscope), digital pathology is most basically defined as viewing renditions of those same images captured by a camera chip composed of an array of light sensing devices, and displayed through a digital monitor. Capture of an image requires a device, such as a camera or scanner, that renders the analog image into an array of “dots” referred to as pixels, each of which has a specific optical density (darkness) and computer-assigned color which is representative of the color that was present in the analog image. Simply put, each pixel assigns a number (or series of numbers) which represents these features and which any computer will recognize and transfer back to the original optical density and color when the image is output onto a viewing device ( Fig. 34-2 ). The array of pixels can then be displayed on a digital output device (e.g., monitor) to give a rendition of the original analog image. Widely available home-use digital cameras are a simple example of this functionality, and the great utility of such devices goes a long way toward explaining the rapid demise of film-based imaging technology. Digital pictures can be easily stored in all sorts of devices, they can be rapidly transmitted to any location, they can be acquired and deleted quickly, and require no post-acquisition processing. This functionality is not available in film-based systems. The resolution of the image is determined by the number and size of the pixels in the array, and, as any photographer will know, the more pixels in the image, the larger and more finely resolved the image can be without distortion (so-called “pixelation”). Resolution comes with costs, including higher memory requirements to store the data, and the need for greater bandwidth capacity to transfer those images to remote sites over networks, such as the internet. Thus, as cameras have become more powerful (meaning that they can capture images using larger and denser pixel arrays), the ability to practically digitize pathology specimens at high enough resolutions to approach the functionality of an analog image is now a reality. In addition, with the development of high-resolution scanners, which are essentially cameras that digitize images across large arrays and combine those images (so-called “stitching”) into yet larger arrays, it is now possible to fabricate final image files that recapitulate entire pathology specimens (so-called “whole slide images” or “virtual slides”). But despite the fact that whole slide images (WSIs) are the medium that has garnered the most attention, other digital methods which are less expensive and less complex (e.g., digital screen sharing) have very useful (and sometimes more functional) applications in actual practice settings. A number of excellent review articles on digital cytology have been recently published and the reader is encouraged to review these for further specific detail.
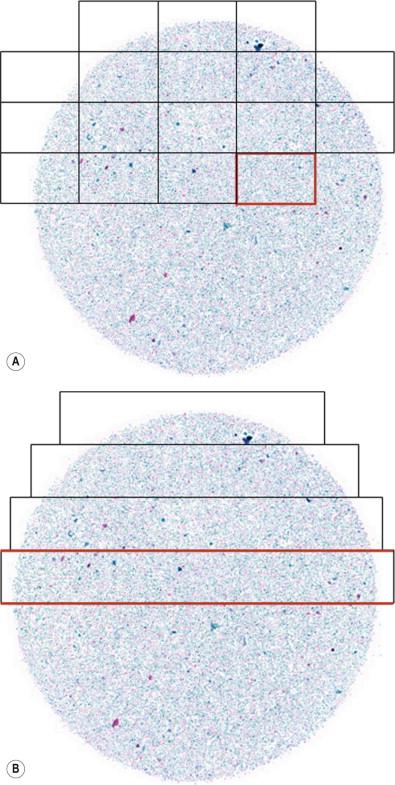
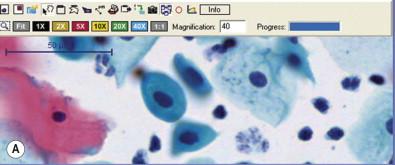
As noted above, digital images are obtained via cameras having arrays of detectors. For the purposes of digital pathology, there are two major classes of acquisition camera devices. The first is a standard digital microscopy or gross image camera. These can range from “smartphone” cameras to dedicated microscope or gross stand-mounted devices. Smartphone devices have been attractive in a variety of telemedicine settings in recent years, with applications for connecting healthcare workers to remote sites for the photographic evaluation of wounds, dermatologic conditions, and, more recently, microscopic detection of blood-borne infections such as malaria. Microscopic attachments to smartphones have allowed on-site and remote interpretations (via wireless networks) to be made where microscopes are not available. In addition, standard digital camera setups allow sharing of static images for diagnostic and educational purposes. The limitation of the standard digital camera is that it cannot capture an entire specimen at high resolution, but is limited to only a small portion of the specimen. The second digital pathology format is the so-called “whole slide image” or virtual slide. WSIs are nothing more than very large static images acquired by specialized instruments that digitize an entire specimen at high resolution. WSIs are made using scanning devices with software that allows for the seamless piecing together of many smaller images. The most important technology jump between standard cameras and whole slide imaging scanners is not the camera itself, but the “stitching” software that assembles the entire specimen image ( Fig. 34-3 ). WSIs can then be viewed on monitors allowing for movement of the image and magnification changes in a manner analogous to the movement of a slide on a microscope stage, hence the name “virtual slide.”
There are a number of manufacturers of WSI scanners on the market at present. The reader is urged to review options in real time as the field is moving fast. The general types and features of scanners will be presented without reference to specific manufacturers or devices, as it is likely that any particular device will be outdated quickly.
Scanners range from relatively simple devices that scan single slides; to intermediate-level devices that will automatically scan a small number of slides (e.g., 5–15 slides); to sophisticated devices with robotic loading of many (>300) slides, which can be processed automatically ( Fig. 34-4 ). Slide scanning speeds vary among the devices, ranging from many minutes per case down to less than 2 minutes. Scanning speed is highly dependent on not only the particular apparatus, but also the settings used for each particular slide scan. Automatic focus features, user-defined focus points, multiple plane scanning, and image compression features will all have an impact on the time to scan. In addition, post-scanning image processing (stitching and image viewing formatting) adds further time before a scanned WSI is available for review. The magnification at which a slide is scanned is also important, not only for speed but also for final viewing resolution. Most current scanners use a ×20 magnification objective lens for fastest scanning, but it is wise to remember that not all ×20 objectives are created equally. The numerical aperture of the lens used also has an effect on the final image created, and hence some ×20 scans are not as high resolution as others. The newest scanners also have the capability of scanning using ×40 or even higher magnification objective lenses. These create images with higher resolution which can therefore be viewed at higher final magnifications. Higher magnification scanning takes longer (all other things being equal) as each individual field of view is smaller. The trend in the industry is to now develop faster high-magnification scanners because applications, particularly diagnostic, require higher resolutions for confident interpretations (see below). In addition, some new software applications allow for the making of WSIs without the need for scanners. These devices are essentially small cameras that use software manipulations to stitch images together as a slide is moved manually across the stage of a standard microscope. When the camera has imaged the entire specimen, the final stitched image is a WSI. Because of its low cost, such technology has the potential to make WSIs available to a wider audience – particularly for applications requiring the scanning of only a selected small numbers of slides.

There are three basic digital pathology image transfer methods currently in common use: (1) transfer of small static images taken with standard digital cameras; (2) real-time (or streaming) image transfer (also using images taken with standard digital cameras); or (3) the transfer of WSIs.
Static image transfer has the advantage of ease of use and relative low cost. This process requires only a camera, computer, and network connection to operate. Image size (as compared to WSIs) is small and bandwidth requirements are low. However, static images show only a limited portion of the entire specimen. Therefore, remote observers will only see what the originator has imaged. This situation may be perfectly acceptable for education and testing purposes, but creates a bias which may be detrimental in the particular circumstance of telediagnosis/consultation where the specimen is an unknown, for which a diagnostic opinion is being sought. In “simple” cases, this may not be a significant issue, but in difficult or true “consultation grade” cases, this bias could potentially skew the opinion rendered. The lack of any ability to focus, change magnifications, or otherwise manipulate the image can also be detrimental to optimal interpretation. One study used video image capture as a method of solving the focusing problem with static imagery. The authors report that using a specialized digital camera capable of capturing short video clips of manual focusing (a “z-axis video”) improved diagnostic ability compared with non-focused still images. When the review has been completed, the images can be permanently archived.
Real-time image transmission or streaming of a digital image solves some of the problems of static image transfer, but at a cost. In this process, the specimen is imaged with a camera attached to a microscope at the originating site, and a continuously updated digital image is streamed over the network to the remote site. The microscope can be either locally or remotely controlled (the latter via a robotic interface) with, in the latter case, commands sent over the interface to move the slide and change the focus or magnification. Therefore, the entire slide can be visualized in a manner entirely analogous to primary microscopy, thus eliminating the bias of static imaging. However, this process can be exceedingly cumbersome, as the manipulation of the remote microscope can be tedious and slow. Real-time systems work best for histopathology applications because much of the work is done at lower magnifications, allowing greater areas of the slide to be viewed in a smaller amount of time. Real-time imaging is least useful for applications requiring high-magnification searching or screening, hence its use for the primary evaluation of cytologic material is most limited. Real-time high-resolution image transfer requires high bandwidth and can easily overload network capacity. Lower resolution systems require less bandwidth, but suffer from lack of fine detail. When the real-time review is completed, no stored image or record of the review exists, unless static images are recorded during the session. Real-time remotely controlled robotic devices are expensive, however use of locally controlled real-time image systems are inexpensive and can be efficiently deployed. One variant of real-time imaging which has significant promise for telepathology is the use of “screen-sharing” software to view images. In this method, remote sites use a standard digital camera attached to a microscope to bring images to their own computer screens. Then screen-sharing programs (Web-X programs) allow other internet-connected users to view that screen in real time. This application has been successfully used for remote frozen section and rapid cytology interpretations, and remote cervical cytology pathologist reviews in international initiatives where pathologists are not readily available on site ( Fig. 34-5 ). Again, similar to static image transfer, the receiving viewer sees only images selected by the sender, creating a bias; there is no remote control of specimen viewing with this method. The real-time nature of this process allows, however, for communication and hence interaction with the remote site, maintaining the potential for manipulation of the specimen.
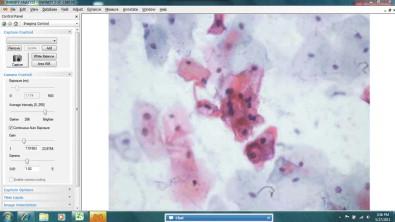
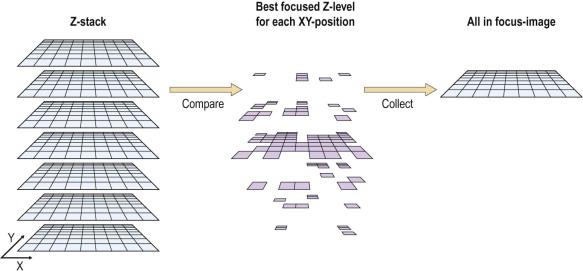
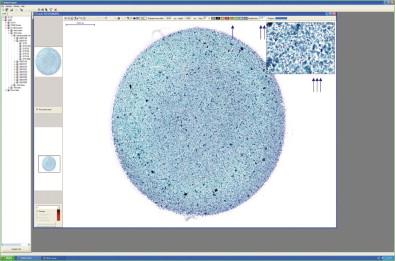
Whole slide imaging maintains the relative simplicity of static image transfer, but eliminates its bias because the entire specimen is available for review. This process creates a very large file size for each image (WSIs can be many hundreds of megabytes of data each), and hence the memory required to store, and the bandwidth required to transfer, such images is great. At present, the WSI scanning process is lengthy, particularly for cytology specimens where high resolution and multiplane scanning may be necessary. A single plane ×20 scan can take up to 5 minutes depending on the area of the specimen, and with the higher resolution scanning that will optimize the high-magnification (×40 objective scanning) image for cytology specimens, scanning can take 30–40 minutes. Faster scanner mechanics or multiple array objectives will undoubtedly mitigate this scanning time process in the future. As with static imaging, WSIs share the lack of intrinsic focusing capability. For cytologic material, which is naturally displayed in a multiplanar fashion, this can be a detriment, although there are methods to mitigate the 3-dimensional aspects of cytologic WSIs. Multiplane scanning of a specimen (so-called “z-stacking”) allows for capture of many focal planes, with the ability of the viewer to “focus” up and down through the planes to find optimally focused cells in each area of the specimen ( Fig. 34-6 ). Z-stacking requires the storage of file sizes that are multiples of the already large single plane WSIs. Systems currently in use to transfer z-stacks over networks can be very slow and can therefore lead to user fatigue and frustration. Going forward, these downside issues of z-stacking may be viewed as “engineering” problems that will be solved with faster scanners, computers and networks; and with ever-increasing storage capacities. Alternative solutions to z-stacking for the 3-dimensionality of cytologic specimens have been achieved using devices that allow for multiplane scanning with integration of the best focused image at each tile into a final single plane file ( Fig. 34-6 ). Scanning time is not improved over z-stacking, as each plane still needs to be scanned individually, but the final integrated file uses the memory and transfer requirements of a single plane scan. Focus of the WSI has been shown to be significantly improved using this method ( Fig. 34-7 ). Interestingly, a study aimed at defining differences in performance of observers using 2-dimensional vs. 3-dimensional cytology images showed no difference in diagnostic accuracy; however, it did highlight a higher level of “uncertainty and frustration” of the observers who were unable to resolve out of focus cells on the 2-dimensional WSIs. At present, the major limiting factor in the use of WSIs is the high cost of the scanning devices. Bandwidth requirements for WSI transfer may not be onerous because modern transfer techniques do not always require transfer of the entire image file. The complete image file can reside on the remote server and only the pieces of the image being queried at any moment can be transferred. Newer software also increases the speed of viewing by transferring the surrounding areas in anticipation of the observer's next move. An example of a typical viewing station for a WSI is illustrated in Fig. 34-8 . Table 34-1 shows a summary of the key features of the three methods of digital image transfer described above.
| Feature | Static | Real Time | WSI |
|---|---|---|---|
| Speed of acquisition | Quick | Medium | Slow |
| Speed of viewing | Quick | Slow | Medium |
| Focus capability | None | Focusable | May be either |
| Amount of specimen | Portion | Whole | Whole |
| Selection bias | Yes | No bias | No bias |
| Magnification | High | High | Dependent on acquisition magnification |
| Permanent record | Yes | No | Yes |
| Annotation | Yes | Possible but not routine | Yes |
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here