Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
We thank Álvaro Macías Martínez, Francisco Miguel Cruz Uréndez, Ana Isabel Moreno Manuel, Lilian Karina Gutiérrez Espinosa de los Monteros, and Isabel Martínez Carrascoso for their helpful discussions and critical review of the manuscript. J.J. has received funding from “la Caixa” Banking Foundation under the project code HR18-00304. Support also came from a Severo Ochoa CNIC Intramural Project (Expediente 12-2016 IGP) to J.J. The CNIC is supported by Instituto de Salud Carlos III (ISCIII), Ministerio de Ciencia e Innovación, and Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
The emergence of human-induced pluripotent stem cell (hiPSC) technology 14 years ago substantially modified the way biomedical research is conducted. hiPSCs are becoming an essential tool in all fields of biomedical science and have opened new horizons for cardiac researchers. Differentiation of hiPSCs into cardiovascular lineages, including cardiomyocytes (CMs), smooth muscle, endothelial cells (ECs), and fibroblasts, is now routine and a highly efficient process thanks to the hard work of many labs worldwide. This is important for an organ with essentially no regenerative capacity like the mammalian heart. Furthermore, unlimited numbers of hiPSC-derived CMs (hiPSC-CMs) can be generated, and the possibilities for personalized medicine are highly promising.
In 2006, Takahashi and Yamanaka first reported their ability to reprogram mouse embryonic or adult fibroblasts into iPSCs by introducing the four Yamanaka transcription factors, Oct3/4, Sox2, c-Myc, and Klf4, under culture conditions ( Fig. 59.1 ). This important scientific discovery made it possible one year later to reprogram human somatic cells into hiPSCs. It then became feasible to generate patient-specific hiPSCs, and subsequently derive hiPSCs into hiPSC-CMs and other cell types. ,
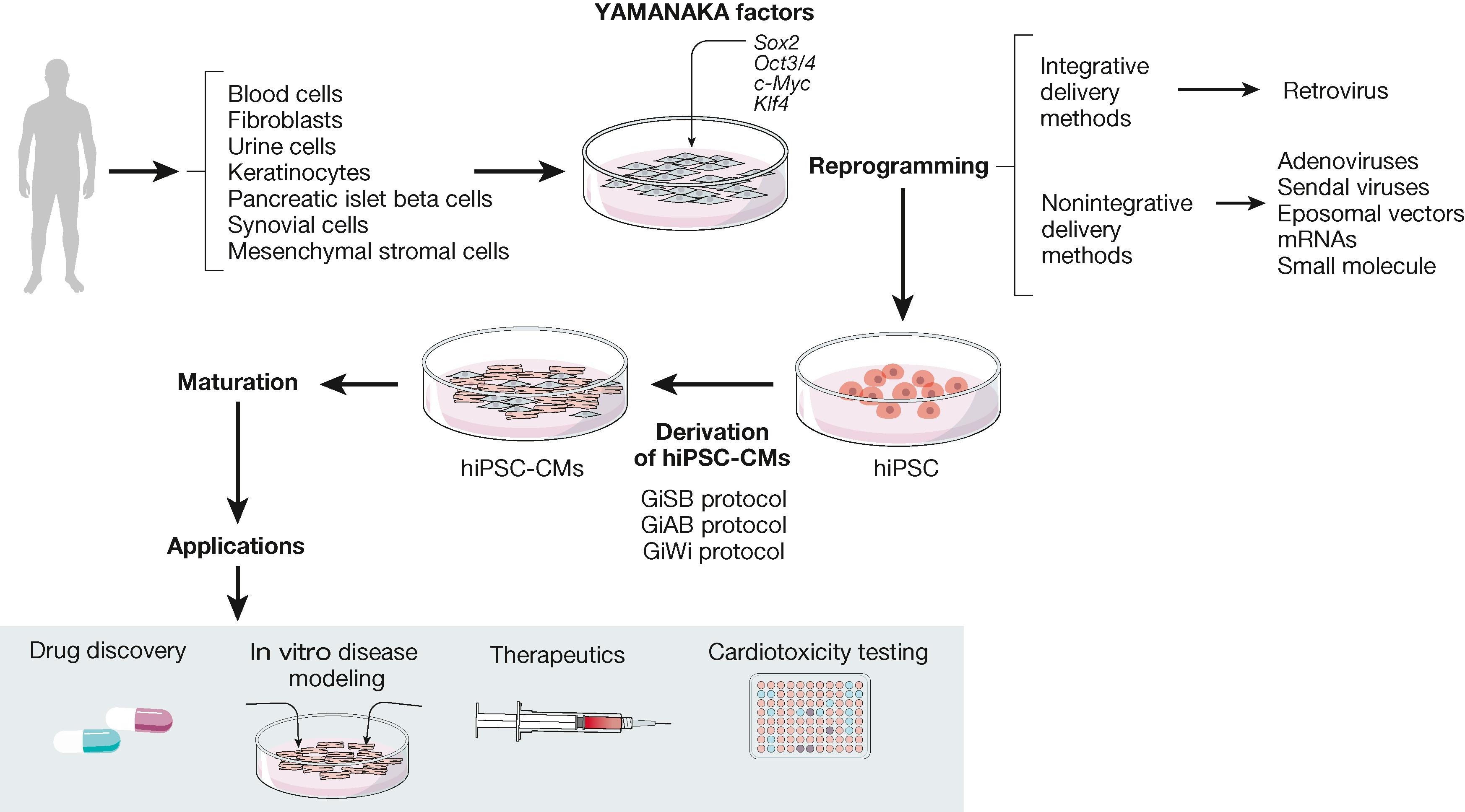
Today millions of hiPSC-CMs can be generated, offering potentially exciting platforms to investigate cardiac arrhythmia mechanisms, and for preclinical cardiotoxicity testing, proarrhythmia screening, drug discovery, and regenerative medicine (see Fig. 59.1 ). However, for the most part hiPSC-CMs generated display immature embryonic-like functional and structural characteristics that limit their utility for cardiac research, regeneration, or cardiotoxicity testing. Typically, single hiPSC-CMs, hiPSC-CMs monolayers, organoids, and bioengineered constructs reported in the literature have immature properties in terms of gene expression, metabolism, myofibrillar structure, contractility, ion channel function, excitability, action potential profile, and conduction velocity, which make them differ substantially from adult cardiac myocytes. Therefore despite great enthusiasm for hiPSC-CMs in the cardiovascular field, many questions remain about the relevance of findings derived from their use. Many laboratories are working to improve the maturity of hiPSC-CMs using a variety of approaches, including prolonged periods of culture; culturing on soft surfaces; coculture with other cell types; chemical, electrical, or mechanical stimulation; three-dimensional (3D) cell culture conditions; and combinations thereof. However, none has proven to yield the archetype adult-like hiPSC-CM. In this review, we briefly evaluate some of the approaches that are used currently to reprogram human somatic cells into hiPSCs. We then focus on the increasingly successful protocols that have been explored for the derivation, purification, and maturation toward the adult-like CM phenotype. Although the ideal conditions for hiPSC-CM maturation have yet to be achieved, we acknowledge the contribution of many scientists and their laboratories toward achieving that significant goal and recognize that this field is moving forward at an accelerating speed. We also hope this chapter will inspire other scientists to further help define the precise cellular and molecular mechanisms at play during the hiPSC-CMs maturation process toward improving the capacity of these wonderful cells to fulfill their promise as a superlative human ex vivo research tool.
hiPSCs are generated by reprogramming different types of somatic cells, but fibroblasts are the most often used primary somatic cell type for this purpose. Fibroblasts offer many advantages; they can be easily isolated from skin biopsies, then cultured, passaged, and cryopreserved, and are uncomplicated with respect to nutritional requirements and viability in culture. However, skin biopsies are invasive, and many patients and their families are reluctant to undergo the procedure. Therefore a number of investigators have shifted to reprogramming other human somatic cell types, including cells isolated from blood samples or urine; keratinocytes from plucked human hair; or even pancreatic islet beta cells, synovial cells, or mesenchymal stromal cells from wisdom teeth of healthy donors; or patients carrying genetic mutations (see Fig. 59.1 ). To reprogram the cells one can use a retrovirus, or better yet nonintegrative delivery methods, such as adenoviruses. Sendai viruses, episomal vectors, mRNAs, and small molecules are also being used. Moreover, cell lines generated from patients, using genome editing tools such as zinc-finger nucleases (ZFNs), transcription activator–like effector nuclease (TALENs), clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short palindromic repeat–associated 9 (CRISPR/Cas9), or single nucleotide polymorphisms (SNPs) allow the generation of isogenic control cell lines through replacement of the mutated allele by the healthy version. Pluripotency is usually confirmed by immunofluorescence analysis of pluripotency markers (e.g., Oct-4, Sox2, Nanog, and Kfl4 ), embryoid body differentiation to demonstrate spontaneous transformation of hiPSC into three germ layers (ectoderm, mesoderm, and endoderm), and karyotyping to identify any chromosomal abnormalities.
Loss of genetic integrity may be a source of variability that may invalidate the use of hiPSCs in disease modeling, drug testing, or reprogramming. In addition, certain genetic variants could be associated with increased risk of cancer or dysfunction when using these cells for regenerative therapeutic interventions. Therefore quality control involving confirmation of normal chromosomal arrangement and a detailed characterization of genetic differences between donor and derived cells are essential to ensure validity and safety.
Derivation of hiPSCs into hiPSC-CMs involves several signaling pathways for mesoderm induction and cardiac specification. , Several protocols, including permutations of growth factors, cytokines, small molecules, and culture medium, might be used to reach an efficient level of hiPSC-CMs production. , , In fact, these molecules present a different association with the three germ layers. Notably, the bone morphogenetic protein (BMP) family is expressed in the endoderm, which is in direct contact with the cardiac mesoderm, and is also linked to the ectoderm and extraembryonic tissues, facilitating cardiac development.
Creating increasingly robust and cost-effective protocols to differentiate hiPSCs into CM-like cells has greatly increased their use. One such protocol is based on the use of growth factors of the transforming growth factor-β (TGF-β) superfamily (GiAB protocol) ( Fig. 59.2 ). The approach uses the glycogen synthase kinase-3 (Gsk3) inhibitor followed by Activin A and BMP4 in RPMI/B27-insulin medium. Another protocol is based on the activation of canonical Wnt signaling followed by shRNA of β-catenin expression, in RPMI/B27-insulin (GiSB protocol). The latter protocol requires a genetic modification of the hiPSC line under the activity of a fluorescent marker or a cardiac-specific promoter in the hiPSC line. The so-called monolayer-based cardiac-directed differentiation protocol is highly efficient, reproducible, and routinely yields approximately 60% to 70% cardiac troponin T–positive (cTnT+) CMs by day 15 of the procedure (see Fig. 59.2 ). , It uses Wnt signaling inhibitors instead of β-catenin shRNA and obviates modification of the hiPSC line. , , This approach is based on small molecule inhibition of Wnt signaling by the Gsk-3 inhibitor (CHIR99021) followed by inhibition of Wnt signaling via the inhibitor of Wnt production 2 (IWP2) or inhibitor of Wnt production 4 (IWP4) using RPMI/B27-insulin (GiWi protocol) (see Fig. 59.2 ). However, although the process is critically dependent on the regulation of the Wnt signaling pathway, the precise mechanism through which the Wnt inhibitor acts on cardiac mesoderm during heart induction has not been established. In addition, questions remain about specific dose and timing needed and interaction among the inducers that regulate Wnt in mesendoderm specification to cardiac mesoderm.
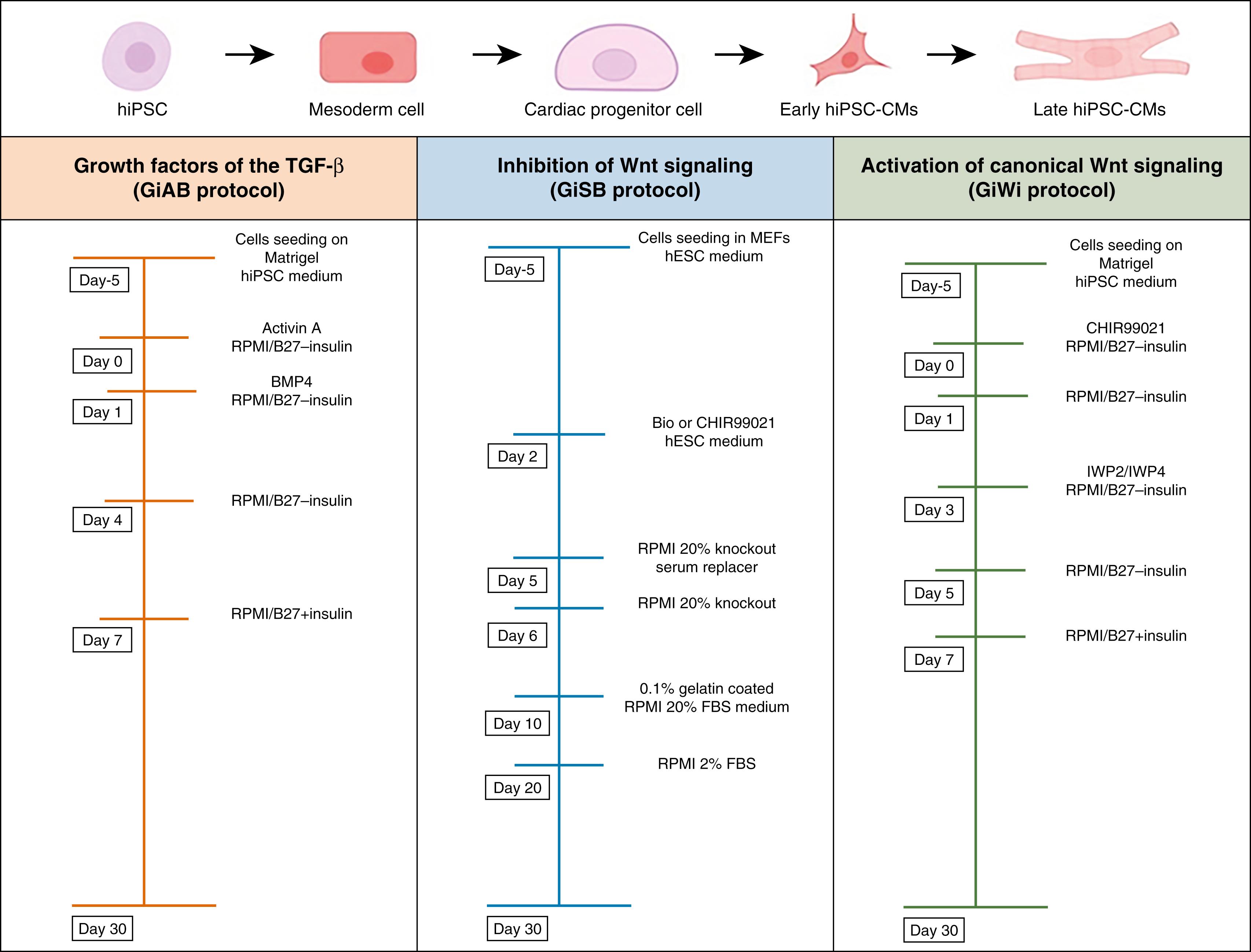
The extracellular matrix (ECM) plays an important role in stem cell fate decisions, cell development, and cardiogenesis. , , In addition to providing structural support for the developing myocardium, the ECM also contains important signaling molecules. Zhang and coworkers developed the matrix sandwich protocol by culturing pluripotent stem cells as monolayers on Matrigel, an ECM complex protein mixture, subsequently overlayed with Matrigel (making a “Matrigel sandwich”). The approach promotes an epithelial-to-mesenchymal transition with the generation of N-cadherin-positive mesenchymal cells. Sequential application of Activin A, BMP4, RPMI/B27-insulin, and RPMI/B27 + insulin to the mixture generated up to 11 CMs per input stem cells with 98% purity from multiple stem lines. Allowing the resulting CMs to progressively mature over 30 days in culture establishes the conditions for structure and electrical function typical of embryonic CMs, including myofiber assembly; mitotic activity; and nodal, atrial, and ventricular CMs. Optical mapping of CM monolayers derived from this protocol usually demonstrates that they are electrically coupled, propagate impulses, and can be induced to generate focal and reentrant arrhythmias. The development of the matrix sandwich protocol demonstrated the critical role of the ECM for highly efficient cardiac-directed differentiation of human pluripotent stem cells.
In every case, the final product of each of the previously mentioned differentiation protocols is a heterogeneous population of atrial-, nodal-, or ventricular-like CMs, and non-CMs. In fact, even the most efficient cardiac-directed differentiation protocols generate CMs and other cell types like fibroblasts and ECs. In many cases, the experiment requires purified CMs, through the removal of the non-CM population from the differentiated pool. There are traditional methods for purification, such as density gradient centrifugation, and genetic selection of cells with markers such as NKX2-5 , MYH6 , MLC2V , and ISL1 . Metabolic stress selection, fluorescence-activated cell sorting (FACS), or magnetic bead sorting can also be used based on specific markers expressed in CMs, such as EMILIN2 , SIRPA , , and VCAM . ,
Metabolic stress selection is currently one of the most efficient and widely used hiPSC-CMs purification approaches. CMs, unlike other cells, can use lactate in the absence of glucose. Thus in a glucose-free and lactate-rich environment other cells die while the CMs survive. , Advantages of this approach include cost-effectiveness, simple bulk purification, low user-to-user variability, and high efficiency. However, the glucose-free and lactate-rich purification medium simulates metabolic conditions related to myocardial ischemia, which must be considered when using hiPSC-CMs purified by this method.
Antibody-based selection relying on magnetic microbeads to separate cells using unique extracellular receptors is currently used to generate cell therapy to treat ischemic heart disease. , This technology is called magnetic cell sorting (MACS) and is approved for the Good Manufacturing Practice (GMP) generation of clinical-grade cells to be used as therapies. A methodologically novel, clinically relevant workflow for hiPSC-CM sorting was validated by Todd Herron at the University of Michigan in collaboration with Miltenyi Biotec. The approach enables targeting of the nonmyocyte population using a magnetically labeled antibody to trap the nonmyocytes in the magnetic column while the hiPSC-CMs flow through and the resulting cell population is hiPSC-CM enriched. The MACS and metabolic stress selection approaches are equally well suited to generate highly purified hiPSC-CMs populations, although the former yields hiPSC-CMs with better electrophysiologic characteristics than the latter. The new MACS approach is also a significant improvement over using SIRPA2a expression to positively select for hiPSC-CMs, which we have used in previous publications. , Using MACS one can isolate hiPSC-CMs from a starting population of just 57.7% cTnT+ cells and obtain enriched hiPSC-CMs with 97.1% purity without significant loss of CMs. Confocal imaging and immunostaining for α-actinin verify the purification method and show the formation of electrically coupled two-dimensional (2D) microtissues.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here