Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
![]() Additional content is available online at Elsevier eBooks for Practicing Clinicians
Additional content is available online at Elsevier eBooks for Practicing Clinicians
Clinicians use biomarkers daily in the practice of cardiovascular medicine. Moreover, the use of biomarkers can continue to improve physicians’ ability to provide clinically effective and cost-effective cardiovascular medicine in the years ahead. Appropriate risk stratification and targeting of therapies should not only help improve patient outcomes but also assist in responding to the urgent need to “bend the cost curve” of medical care. In particular, excessive use of imaging biomarkers increases the cost of medical care and can jeopardize patient outcomes (e.g., from radiation exposure or complications of administering contrast material or investigating incidental findings). Inappropriate use or interpretation of blood biomarkers (e.g., cardiac troponin levels) can lead to unnecessary hospitalization or procedures as well.
Despite the current usefulness of biomarkers, their future promise, and the critical need to use them appropriately, much misunderstanding still surrounds their current clinical application. In addition, contemporary technologies can greatly expand the gamut of biomarkers relevant to cardiovascular practice. Emerging genetic, proteomic, metabolomic, and molecular imaging strategies will surely transform the landscape of cardiovascular biomarkers (see also Chapter 7, Chapter 8, Chapter 25 ).
This chapter provides a primer on cardiovascular biomarkers by defining terms and discussing how the application of biomarkers can assist in clinical care. The literature abounds with descriptions of biomarkers offered to apply to various clinical situations. Advances in cardiovascular biology and the application of novel technologies have identified a plethora of novel cardiovascular biomarkers of potential clinical usefulness—begging the question of whether a novel biomarker adds value to existing and often better-validated biomarkers. Thus clinicians need tools to evaluate these emerging biomarkers, to discern which may actually elevate clinical practice and improve patient outcomes. To help the reader in this regard, we also provide a guide to the rigorous evaluation of the clinical utility of biomarkers. Chapter 8 explicates the application of proteomics and metabolomics to discover novel biomarkers.
For regulatory purposes, the U.S. Food and Drug Administration (FDA) first defined a biomarker in 1992 as “a laboratory measure or physical sign that is used in therapeutic trials as a substitute for a clinically meaningful end point that is a direct measure of how a patient feels, functions, or survives and is expected to predict the effect of the therapy.” At that time the FDA considered a surrogate endpoint as “reasonably likely, based on epidemiologic, therapeutic, pathophysiologic, or other evidence, to predict clinical benefit.” The National Institutes of Health (NIH) convened a working group in 1998 that offered some parallel operating definitions to guide the biomarker field ( Table 10.1 ). NIH defined a biologic marker, or biomarker, as “a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.” Thus the NIH definition includes not only soluble biomarkers in circulating blood but also “bedside biomarkers,” such as anthropomorphic variables obtainable with a blood pressure cuff or a tape measure at the point of care.
| Biologic marker (biomarker) A characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. Surrogate endpoint A biomarker intended to substitute for a clinical endpoint. A surrogate endpoint is expected to predict clinical benefit (or harm) or lack of benefit (or harm) based on epidemiologic, therapeutic, pathophysiologic, or other scientific evidence. Clinical endpoint A characteristic or variable that reflects how a patient feels, functions, or survives. |
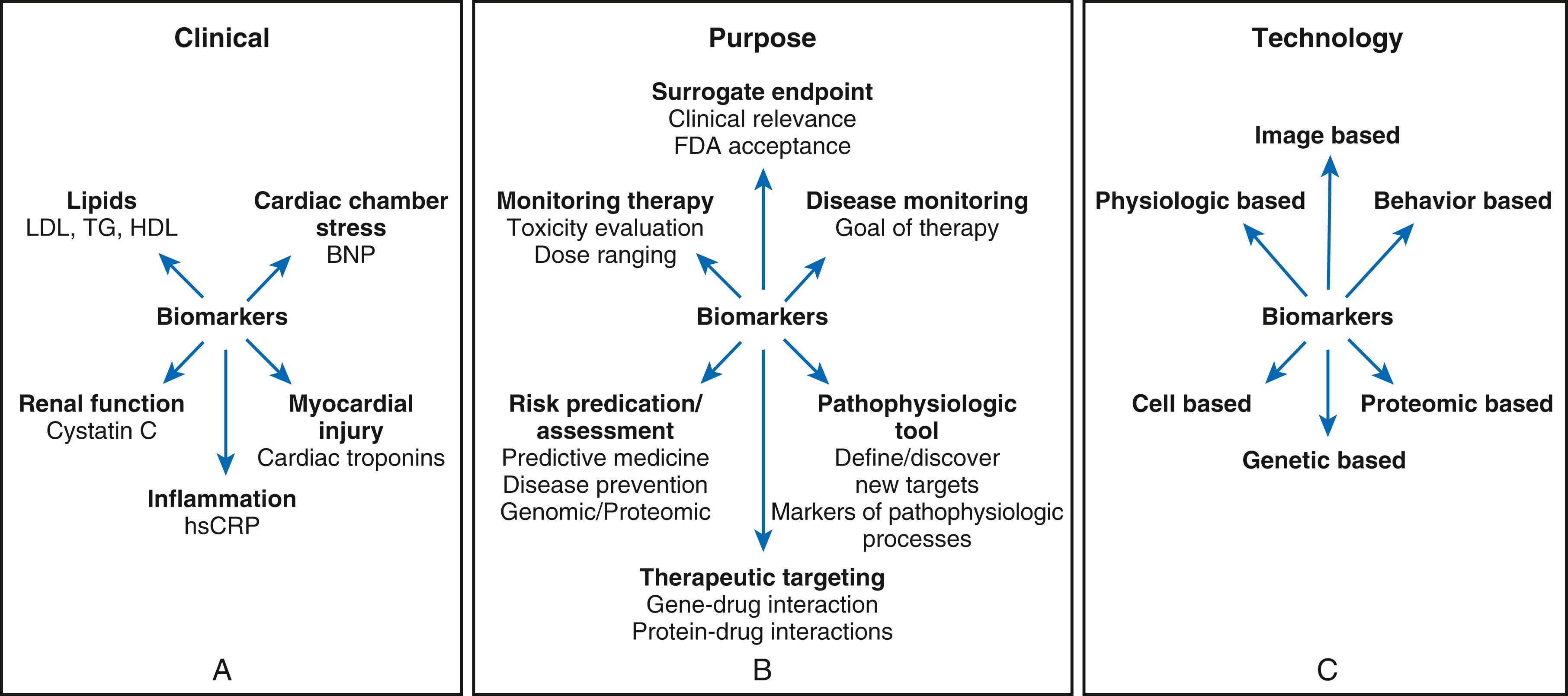
This broad definition encompasses measurements of biomarkers in blood ( Fig. 10.1A ) as well as measurements from imaging studies ( Fig. 10.1B ). Imaging biomarkers can include those derived from classic anatomic approaches. Imaging modalities now offer functional information, such as estimates of ventricular function and myocardial perfusion. Molecular imaging has the potential to target specific molecular processes. A functional classification of biomarkers helps sort through the plethora encountered by the clinician, in that biomarkers can reflect a variety of biologic processes or organs of origin. For example, as shown in Figure 10.1B , to a first approximation, cardiac troponin reflects myocardial injury, brain natriuretic peptide reflects cardiac chamber stretch, C-reactive protein reflects inflammation, and cystatin C and the estimated glomerular filtration rate reflect kidney function.
The NIH working group defined a surrogate endpoint as “a biomarker intended to substitute for a clinical endpoint. A surrogate endpoint is expected to predict clinical benefit (or harm) or lack of benefit (or harm) based on epidemiologic, therapeutic, pathophysiologic, or other scientific evidence.” (Note that the NIH definitions do not include the commonly used term surrogate marker. ) Thus a surrogate endpoint is a biomarker that has been “elevated” to surrogate status. This distinction has particular importance in the regulatory aspects of cardiovascular medicine. For example, the FDA previously accepted a certain degree of reduction in hemoglobin A 1c (HbA 1c ) as a criterion for registration of a novel oral hypoglycemic agent; thus HbA 1c was considered a biomarker accepted as a surrogate endpoint. Current FDA guidance now requires a cardiovascular safety study for the registration of new medications that target diabetes. This policy indicates regulatory doubts about the fidelity of a decrease in HbA 1c as a surrogate endpoint for reduced cardiovascular risk, despite its value as a biomarker of glycemia.
The NIH working group defined a clinical endpoint as “a characteristic or variable that reflects how a patient feels, functions, or survives.” Pivotal or phase III cardiovascular trials aspire to use clinical endpoints so defined. The distinction among biomarkers, surrogate endpoints, and clinical endpoints has crucial implications as practitioners, regulators, and payers increasingly demand evidence of improvements in actual clinical outcomes rather than mere manipulation of biomarkers as a criterion for adoption of a treatment in clinical practice.
In an effort to dispel persistent confusion regarding definitions in the biomarker arena, in 2015 a joint effort of the FDA and NIH developed an online resource denoted BEST (Biomarkers, EndpointS, and other Tools). They formulated a living online resource that furnishes an extended glossary of terms to facilitate standardization ( Table 10.2 ). The BEST definitions overlap with those of the NIH Workshop, but provide more detail of particular relevance to those interested in the regulatory aspects of biomarkers ( Table 10.3 ).
|
| Biologic marker (biomarker): A defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions. Molecular, histologic, radiographic, or physiologic characteristics are types of biomarkers. A biomarker is not an assessment of how an individual feels, functions, or survives. Surrogate endpoint: An endpoint that is used in clinical trials as a substitute for a direct measure of how a patient feels, functions, or survives. A surrogate endpoint does not measure the clinical benefit of primary interest in and of itself, but rather is expected to predict that clinical benefit or harm based on epidemiologic, therapeutic, pathophysiologic, or other scientific evidence. From a U.S. regulatory standpoint, surrogate endpoints and potential surrogate endpoints can be characterized by the level of clinical validation: validated surrogate endpoint, reasonably likely surrogate endpoint, candidate surrogate endpoint. |
Much of the prevailing confusion regarding biomarkers involves framing the question that the clinician wants to answer with the use of a biomarker ( Fig. 10.1C ). We can classify the goals of application of cardiovascular biomarkers into the following rubrics:
Diagnosis. Daily medical practice uses many biomarkers for cardiovascular diagnosis. The current universal definition of myocardial infarction, for example, requires elevation of a biomarker of myocyte injury, such as cardiac-specific isoforms of troponin.
Risk stratification. Familiar examples of biomarkers used in risk stratification in cardiovascular medicine include systolic blood pressure (SBP) and low-density lipoprotein cholesterol (LDL-C). These biomarkers reliably predict future risk for cardiovascular events on a population basis.
Goals for therapy. Contemporary guidelines often specify cutoff points for targets of treatment, for example, a specific level of a biomarker (e.g., SBP, LDL-C) in a particular group of individuals. Practitioners of cardiovascular medicine typically use the biomarker international normalized ratio (INR) to titrate the dosage of warfarin administered to an individual patient. Abundant data support the clinical benefit of maintaining the INR within a certain range in various patient groups, an example of a widely used biomarker that has proven clinical usefulness as a goal for therapy.
Targeting of therapy. In clinical practice, using biomarkers to target therapy has great usefulness and promise as we move toward a more comprehensive “personalized medicine” approach to practice. Examples of biomarkers used to target therapy include troponin measurements to triage patients with acute coronary syndromes for early invasive management and measurement of high-sensitivity C-reactive protein (hsCRP) to allocate statin treatment to individuals without elevated LDL-C.
Drug development, evaluation, and registration. Biomarkers have critical importance in the development of new pharmacologic agents. Biomarkers can provide early signals of efficacy that will help prioritize agents more likely to provide benefit on clinical endpoints in large-scale trials. Clinical trials not infrequently fail because of inappropriate dose selection. Judicious use of biomarkers can help in selecting an appropriate dose of an agent to study in a large endpoint trial. Biomarkers accepted as surrogate endpoints also prove useful to regulatory agencies in granting approval for novel therapies.
Clinical use of cardiovascular biomarkers requires a clear understanding of how they should be used. Many biomarkers provide clinically useful information when measured once at “baseline.” A baseline measurement of high-density lipoprotein cholesterol (HDL-C), for example, correlates inversely with future risk for cardiovascular events. However, serial measurement of biomarkers to document a change does not always guarantee a clinical benefit. In the case of HDL-C, recent large-scale trials that have measured clinical endpoints have cast doubt on the fidelity of a rise in HDL-C as a predictor of clinical benefit (see Chapter 27 ). A single measurement of coronary artery calcium score (CAC) ably predicts future events in statin naïve individuals. Yet, serial measurements of CAC may prove misleading because statin therapy increases calcification, but decreases coronary events.
Biomarkers require rigorous validation before adoption into clinical practice. In cardiovascular medicine, LDL-C has high reliability as a biomarker; it satisfies the modified Koch postulates. LDL levels prospectively predict cardiovascular risk, and decreases in LDL generally correlate with improved outcomes. Not all biomarkers, however, have proved as faithful in predicting clinical events. In the 1960s and 1970s, for example, most of the cardiovascular community considered ventricular premature depolarizations on the electrocardiogram (ECG) as important biomarkers for lethal arrhythmias. Numerous strategies have been aimed at suppressing ventricular ectopy. The Cardiac Arrhythmia Suppression Trial (CAST), however, showed that drugs capable of suppressing ventricular premature depolarizations actually worsened survival. The short-term improvements in indices of cardiac contractility produced by inotropic agents similarly led to worsened clinical outcomes, including increased mortality. These examples illustrate the necessity of rigorous validation of biomarkers before adoption into clinical practice.
Another important consideration in the use of cardiovascular biomarkers involves the question of causality. LDL-C exemplifies a causal biomarker, one that clearly participates in the pathogenesis of atherosclerosis. Its levels prospectively correlate with risk for cardiovascular events and the development of atherosclerotic lesions identified by a variety of imaging modalities. A variety of independent manipulations of LDL-C levels correlate with clinical outcomes. In addition, very strong genetic evidence based on mendelian disorders (e.g., familial hypercholesterolemia) and unbiased genome-wide association scans, as well as mendelian randomization analyses, have established LDL-C as a causal risk factor in atherosclerotic cardiovascular disease and as a generally valid surrogate endpoint offering great value in clinical practice (see Chapter 27 ). Even a well-validated causal biomarker such as LDL-C, however, may mislead under some circumstances. For example, lowering of LDL-C with certain cholesteryl ester transfer protein inhibitors does not appear to lead to clinical benefit. , Other lipid measures such as plasma triglycerides and lipoprotein(a) predict risk, but currently lack definitive evidence that intervention reduces that risk. Ongoing trials of triglyceride reduction and of lipoprotein(a) reduction may move these relationships from casual to causal.
Other biomarkers, although clearly clinically useful, do not participate in the causal pathway for disease. For example, fever has served since antiquity as an important biomarker of infection. Resolution of fever correlates with successful resolution of infectious processes. However, fever does not participate causally in the pathogenesis of infection but merely serves as a biomarker of the host defenses against the infectious process. Sometimes a non-causal downstream biomarker can serve as an effective clinical surrogate for an upstream causal biomarker. For example, the use of hsCRP measurements improves the prediction of cardiovascular risk, and reductions in CRP correlate with clinical benefit in many cases. However, mendelian randomization studies do not support a causal role for CRP itself in the pathogenesis of cardiovascular disease. By contrast, intervention trials demonstrate that upstream drivers of CRP, in particular IL-1b, are indeed in the causal pathway leading to myocardial infarction, stroke, and cardiovascular events.
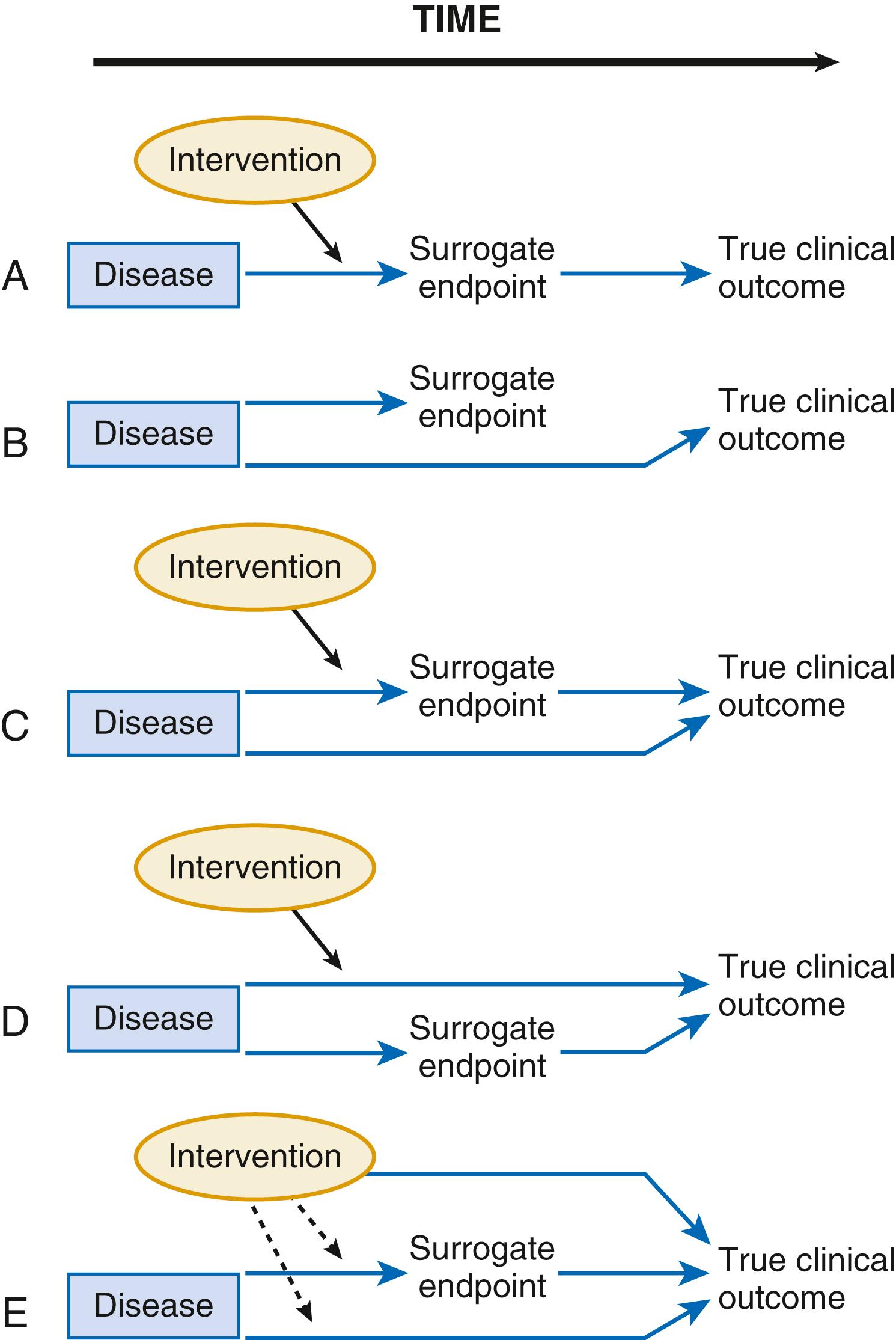
These examples illustrate how a biomarker does not need to reside in the causal pathway of a disease to have clinical usefulness. A clear and early exposition of the uses and pitfalls in the application of biomarkers emerged from the landmark schema of Fleming and DeMets ( Fig. 10.2 ). Biomarkers have the greatest potential for validity when there is one causal pathway and when the effect of intervention on true clinical outcomes is mediated directly through the biomarker surrogate ( Fig. 10.2A ). However, biomarker development can fail when the biomarker is found not to be in the causal pathway, when the biomarker is insensitive to the specific intervention’s effect, or when the intervention of interest has a mechanism of action (or a toxicity) that does not involve the pathway described by the biomarker ( Fig. 10.2B-E ). These examples do not mean that biomarkers lack value; few if any novel biologic fields could develop without biomarker discovery and validation. Still, surrogate endpoints probably will not replace large-scale randomized trials that address whether interventions reduce actual event rates.
The limitations of currently available biomarkers for screening or prognostic use underscore the importance of identifying “uncorrelated” or “orthogonal” biomarkers associated with novel disease pathways (see Fig. 10.1A ). Most current biomarkers have been developed as an extension of targeted physiologic studies investigating known pathways such as tissue injury, inflammation, or hemostasis. By contrast, emerging technologies now enable the systematic, unbiased characterization of variation in proteins and metabolites associated with disease conditions (see Chapter 8 ). The rapid development of polygenic risk scores for cardiovascular disease promises to provide biomarkers that may permit the targeting of therapies particularly in primordial and primary prevention before pathological processes have progressed to the point of altering disease biomarkers (see Chapter 7 .) The applications of machine learning and artificial intelligence (see Chapter 11 ) will doubtless add to the development of novel candidate biomarkers that will require rigorous evaluation for clinical utility as outlined below. The burgeoning field of wearables will provide new inputs into biomarker science as well (see Chapter 12 ). The development of point-of-care technologies will likewise facilitate the clinical application of biomarkers by rendering their use more practical in the field and in urgent situations. Digital technologies, for example, smart phone apps and implanted sensors, will also provide “real world” biomarker input outside of the confines of the traditional medical enterprise.
When considering any biomarker in a clinical setting for risk prediction, physicians should ask two interrelated questions:
Is there clear evidence that the biomarker of interest predicts future cardiovascular events independent of other already measured biomarkers?
Is there clear evidence that patients identified by the biomarker of interest will benefit from a therapy that they otherwise would not have received?
Unless the answer to both these questions is a clear “yes,” measurement of the biomarker will not likely have sufficient usefulness to justify its cost or unintended consequences. Such judgments require clinical expertise and will vary on a case-by-case basis.
Biomarker evaluation also typically involves repeated testing in multiple settings that include varied patient populations and that use different epidemiologic designs. Prospective cohort studies (in which the biomarker or exposure of interest is measured at baseline, when individuals are healthy, and then related to the future development of disease) provide a much stronger form of epidemiologic evidence than do data from retrospective case-control studies (in which the biomarker of interest is measured after the disease is present in the case participants).
After discovery by the technologies described earlier or identification by a candidate approach, a novel biomarker typically requires development in a translational laboratory for refinement of its assay to address issues of interassay and intra-assay variation before any clinical testing begins. Focused studies in specific patient populations typically follow and eventually broaden to encompass the population of greatest clinical interest. Beyond simple reproducibility, biomarkers under development for diagnostic, screening, or predictive purposes require further evaluation with a standard set of performance measures that include sensitivity, specificity, positive and negative predictive value (NPV), discrimination, calibration, reclassification, and tests for external validity.
The validity of a screening or diagnostic test (or one used for prediction) is initially measured by its ability to categorize individuals who have preclinical disease correctly as “test positive” and those without preclinical disease as “test negative.” A simple two-by-two table is typically used to summarize the results of a screening test by dividing those screened into four distinct groups ( Table 10.4 ). In this context, sensitivity and specificity provide fundamental measures of the test’s clinical validity. Sensitivity is the probability of testing positive when the disease is truly present and is defined mathematically as a/(a + c). As sensitivity increases, the number of individuals with disease who are missed by the test decreases, so a test with perfect sensitivity will detect all individuals with disease correctly. In practice, tests with ever-higher sensitivity tend to also classify as “diseased” many individuals who are not actually affected (false positives). Thus the specificity of a test is the probability of screening negative if the disease is truly absent and is defined mathematically as d/(b + d). A test with high specificity will rarely be positive when disease is absent and will therefore lead to a lower proportion of individuals without disease being incorrectly classified as test positive (false positives). A simple way to remember these differences is that sensitivity is “positive in disease,” whereas specificity is “negative in health.”
| Disease Present | Disease Absent | ||
|---|---|---|---|
| Test positive | a | b | a + b |
| Test negative | c | d | c + d |
| Total | a + c | b + d | |
| Sensitivity = a/(a + c) Specificity = d/(b + d) Positive predictive value = a (a + b) Negative predictive value = d/(c + d) |
|||
A perfect test has both very high sensitivity and specificity and thus low false-positive and false-negative classifications. Such test characteristics are rare, however, because there is a trade-off between sensitivity and specificity for almost every screening biomarker, diagnostic, or predictive test in common clinical use. For example, although high LDL-C levels usually serve as a biomarker for atherosclerotic risk, up to half of all incident cardiovascular events occur in those with LDL-C levels well within the normal range, and many events occur even when levels are low. If the diagnostic cutoff criterion for LDL-C is reduced so that more people who actually have high risk for disease will test positive (i.e., increased sensitivity), an immediate consequence of this change will be an increase in the number of people without disease in whom the diagnosis is made incorrectly (i.e., reduced specificity). Conversely, if the criterion for diagnosis or prediction is made more stringent, a greater proportion of those who test negative will actually not have the disease (i.e., improved specificity), but a larger proportion of true cases will be missed (i.e., reduced sensitivity).
In addition to sensitivity and specificity, the performance or yield of a screening, diagnostic, or predictive test also varies depending on the characteristics of the population being evaluated. Positive and NPVs are terms used in epidemiology that refer to measurement of whether an individual actually has (or does not have) a disease, contingent on the result of the screening test itself.
The positive predictive value (PPV) is the probability that a person has the disease of interest, given that the individual tests positive, and is mathematically calculated as PPV = a/(a + b). High PPV can be anticipated when the disease is common in the population being tested. Conversely, the NPV is the probability that an individual is truly disease free, provided that the test has a negative result, and is mathematically calculated as NPV = d/(c + d). High NPV can be anticipated when the disease is rare in the population being tested. Although sensitivity and specificity are largely performance characteristics of the test itself (and thus tend to be fixed values), PPV and NPV depend in part on the population being tested (and thus tend to vary).
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here