Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Molecular mechanisms underlying head and neck squamous cell carcinomas (HNSCCs) are opportunities for targeted therapeutics.
Tobacco and human papilloma virus (HPV), risk factors for HNSCCs, impart genomic patterns; for example, mutation rates are higher for tobacco-associated tumors compared to HPV-associated tumors.
Genetic mutations can be broadly grouped in major pathways: cell cycle (TP53, CDKN2A, MYC), mitogenic (EGFR, RAS, PI3K), differentiation (NOTCH1), and apoptotic.
Epigenetic programming (dysregulation of genes by histone modification, miRNA and DNA methylation) represents a major mechanism involved in cancer development. Targeting the epigenetic modifiers and regulators has become a promising therapeutic avenue.
Tumor microenvironment, which refers to the complex and dynamic interactions between tumor cell and the various surrounding cellular (e.g., carcinoma-associated fibroblast, immune cell system, vascular and lymphatic system) and non-cellular elements (e.g., cytokines, growth factors, extracellular matrix), plays a key role in influencing the behavior of malignant cells.
Tumors can evade the immune system through binding of T-cell receptors such as programmed cell death protein 1 (PD-1). Nivolumab and pembrolizumab, immunotherapies currently used for HNSCCs, block these interactions leading to reactivation of the immune system, thereby mediating recognition and elimination of tumor cells.
Head and neck squamous cell carcinoma (HNSCC) remains a clinical challenge. Overall 5-year survival has improved from 52.7% in 1982–1986 to 65.9% in 2002–2006, with the greatest improvements noted in oropharyngeal carcinomas. While improvement in survival has modestly increased, the challenges in treating HNSCC are unchanged: recurrences, metastases, tumors encroaching upon intricate anatomy, and the inevitable sequelae of treatment.
The molecular biology of HNSCC guides therapeutic approaches. Radiation and chemotherapeutics exploit the high replication rates of tumor cells compared to surrounding normal tissue. The monoclonal antibody cetuximab targets the epidermal growth factor receptor (EGFR), overexpressed in nearly 90% of HNSCC. Finally, novel immunotherapies reactivate the immune system through inhibiting pathways such as programmed death protein 1 (PD-1) and allow the immune system to recognize and eliminate tumor cells.
Over the past decade, research has advanced understanding of the molecular changes in HNSCC. These discoveries include high-throughput genome sequencing leading to a pathway-based organization of mutations, novel mutations such as in NOTCH 1, the interactions with the tumor microenvironment (TME), the evolving story of human papilloma virus (HPV) in HNSCC, and insights on how tumor cells evade the immune system. In the context of abundant research, translating research into clinical care remains the exciting challenge and future of cancer care. This chapter aims to provide updates in the molecular biology of HNSCC.
Since the discovery of the first protooncogene ras and tumor suppressors RB and p53 , our model for cancer mutations and regulation has grown increasingly nuanced. For HNSCC, mathematical models estimated that 6 to 10 accumulated mutations would lead to tumor formation. It is now believed that some mutations can occur simply by chance, known as passenger mutations, while driver mutations impart a growth advantage and therefore are positively selected.
As high-throughput genome sequencing has advanced, new analytic tools and the depth of data acquired have shifted the framework from focusing on individually mutated genes to considering dysregulated pathways, highlighted in landmark papers. In 2011, following whole exome sequencing of HNSCC tumors, two groups independently discovered mutations in NOTCH1 as well as confirmed known mutations including TP53 , PI3K , and HRAS . In 2015, the Cancer Genome Atlas Network assessed 279 HNSCC for genomic mutations, gene copy number variations, and alterations in methylation. Their comprehensive analysis led to broad clustering of mutations by pathways ( Fig. 73.1 ): cell cycle (TP53, CDKN2A, MYC), mitogenic (EGFR, RAS, PI3K), differentiation (NOTCH1), and apoptotic pathways.
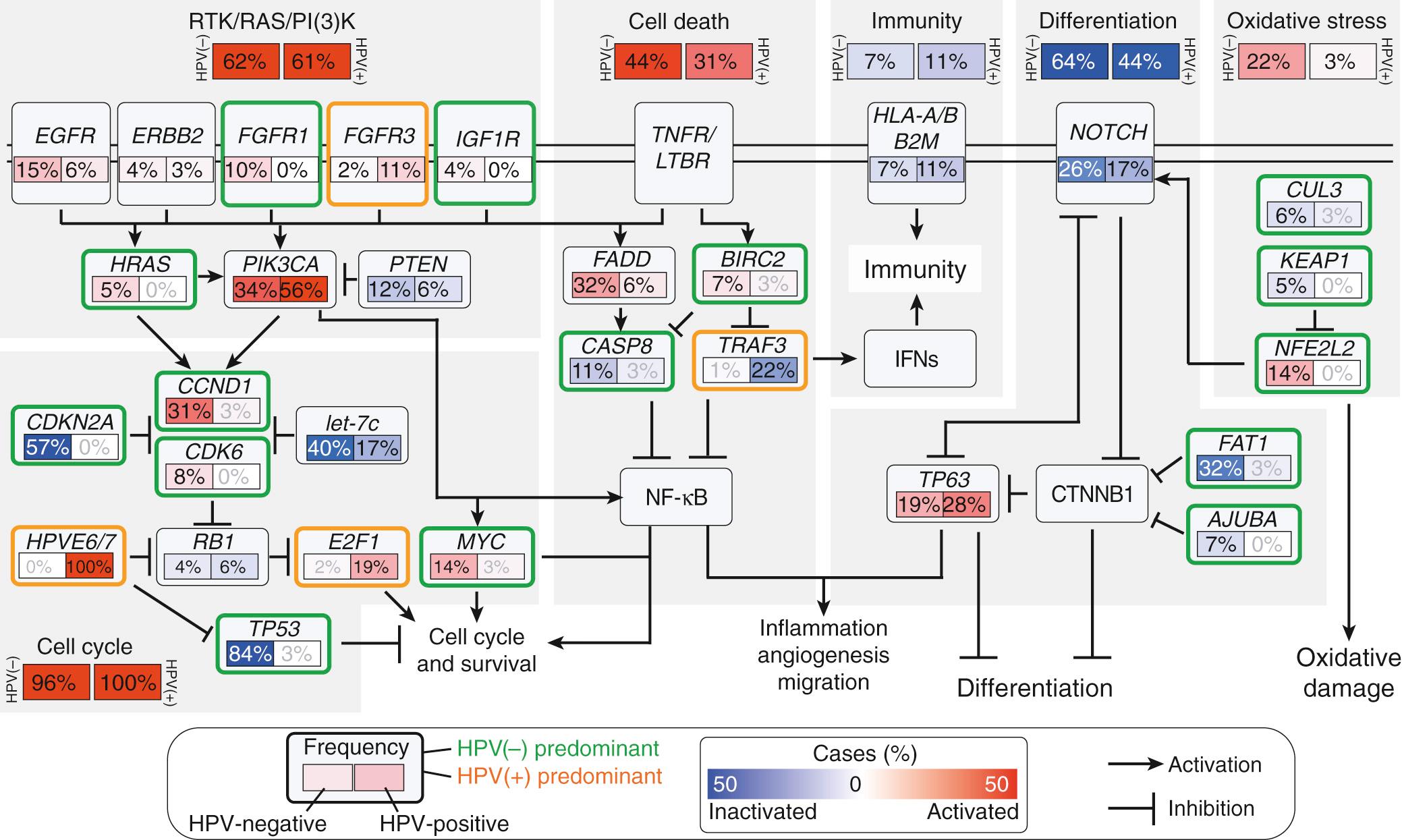
Genomic patterns have emerged from sequencing, such as the heterogeneous nature between and within tumors. In head and neck tumors, intra-tumoral heterogeneity is associated with poorer prognosis. Though not currently integrated into clinical practice, future therapeutic options may eventually alter how these patterns direct clinical care.
Tobacco and HPV (HNSCC risk factors) are also reflected in genomic patterns. Compared to HPV, tobacco exposure is associated with higher mutation rates. Furthermore, Stransky et al. noted that the 4.83 mutations per megabase in tobacco-associated tumors was similar to other tobacco-related tumors such as small cell lung cancer; this mutation rate was twice that of HPV positive tumors. In comparing HPV involvement, HPV positive tumors had fewer mutations than HPV negative tumors.
The cell cycle pathway is the most commonly mutated pathway in HNSCC and targeted by chemotherapy. The Cancer Genome Atlas initiative noted that 96% of HPV negative and 100% of HPV positive tumors harbored cell cycle mutations. This pathway includes established cell cycle regulator p53 (encoded by TP53 ) and cyclin-dependent kinase inhibitor 2A ( CDKN2A ). Furthermore, HPV targets pRB, coded by the tumor suppressor retinoblastoma susceptibility gene 1, and p53 of this pathway (see Fig. 73.2 , see Chapter 74 regarding HPV).
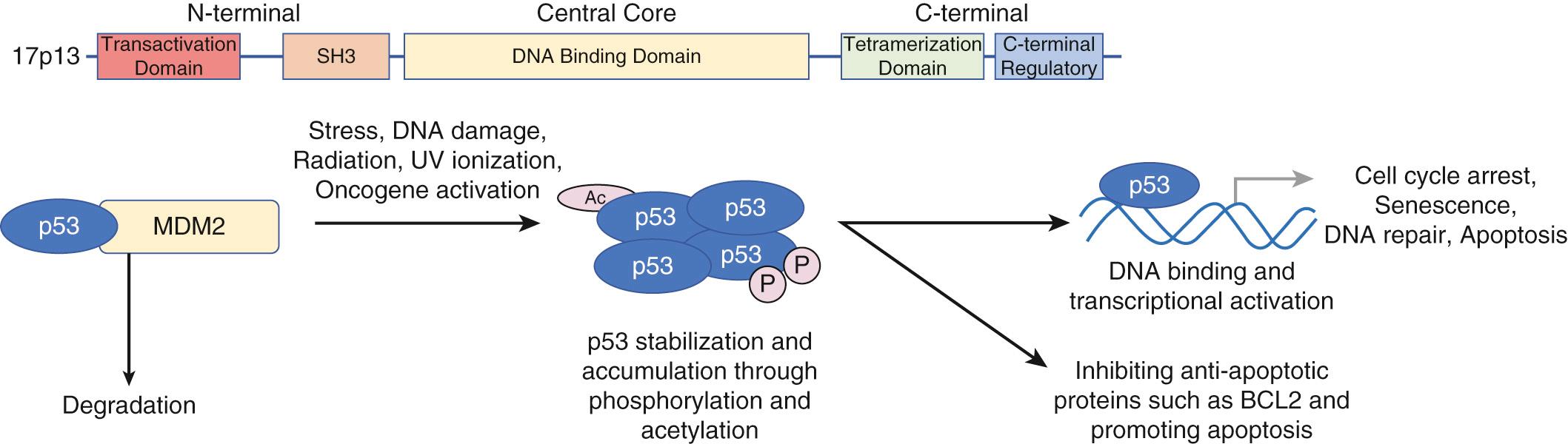
p53, a tumor suppressor on chromosome 17p13, is composed of an N-terminal transcriptional activation domain, central DNA-binding, and C-terminal regulatory and tetramerization domain (see Fig. 73.2 ). MDM2, a negative regulator, normally binds to p53, resulting in protein degradation. However, in the setting of stress and DNA damage caused by factors including radiation and oncogenic activation, upregulation of protein kinases that inhibit MDM2 binding leads to stabilization and activation of p53. This accumulation of p53 causes cell cycle arrest and induces transcription of proteins for DNA repair. If DNA repair is unsuccessful, apoptosis and cellular senescence occur through expression of CDKN1A and BAX. When TP53 is mutated or absent, cell proliferation proceeds despite DNA damage, leading to further accumulation of mutations. An additional level of regulation is through DNA binding. The C terminus region normally inhibits the DNA-binding domain; phosphorylation or acetylation of C terminus residues can promote DNA binding.
TP53 is the most commonly mutated gene in HNSCC. TP53 mutations are early mutations found in premalignant and dysplastic oral cavity lesions, with prevalence increasing with tumor progression. An estimated 47% to 72% of HNSCC harbored TP53 mutations, with nearly 50% to 63% of these being missense mutations. Tobacco exposure was associated with increased TP53 mutations. In contrast, there appears to be an inverse relationship between TP53 mutations and HPV positivity. Clinically, mutations have been associated with decreased overall survival, increased local regional recurrence, and decreased response to chemotherapy and radiation. In a prospective study of 420 patients by Poeta et al., HNSCC patients with p53 mutations were noted to have a decreased 5-year overall survival of 40.7% compared to 54.8% for patients with wild-type TP53 ( P = .009); furthermore, among the TP53 mutations, patients with more disruptive mutations predicted to perturb the DNA binding domain or mutations resulting in stop codons had a median survival of 2.0 years compared to 3.9 years for patients with remaining types of mutations as detected by the Affymetrix gene chip and liquid chromatography.
The cyclin-dependent kinase inhibitor 2A ( CDKN2A ) gene resides on chromosome 9p21 and codes for p14 ARF and p16 INK4A , which regulate p53 ( Fig. 73.3 ). Both gene products inactivate proteins, which prevent phosphorylation of retinoblastoma, thereby inhibiting transcription factor E2F. This leads to cell cycle arrest by preventing progression to the S phase. p14 ARF specifically inhibits MDM2, which stabilizes p53, thereby allowing cell cycle and DNA repair. p16 binds to cyclin-dependent kinases CDK4 and CDK6, leading to formation of the Rb-E2F complex, which results in G1 arrest. Phosphorylation of CDK4 and CDK6 releases E2F; this in turn activates genes for cellular replication and progression to S phase.
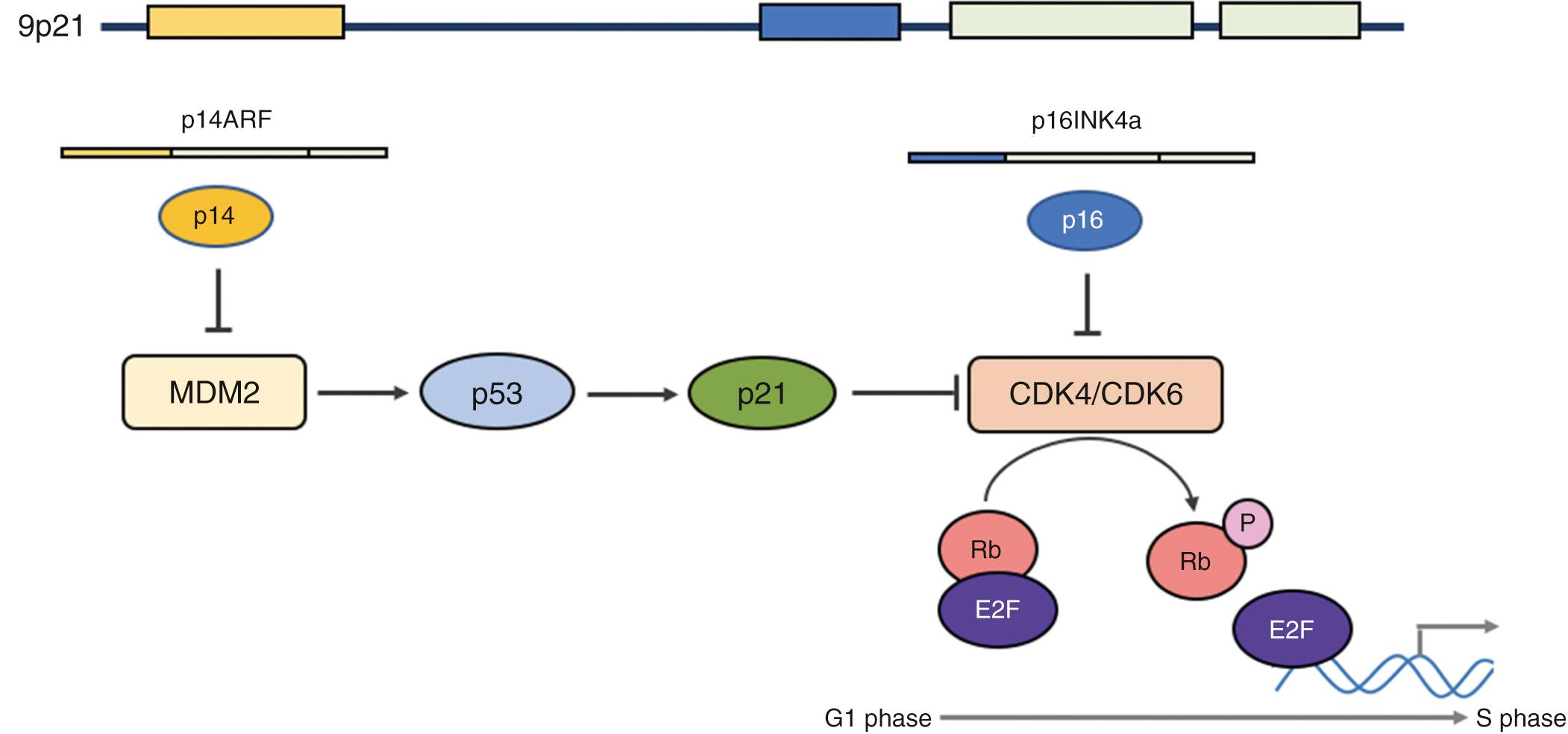
CDKN2A is a tumor suppressor gene (TSG) somatically mutated in 9% to 12% of HNSCC and inactivated in an estimated 60% to 90% of HNSCC through homozygous deletions and epigenetic silencing through promoter methylation. More mutations are noted in HPV negative tumors. In HPV positive tumors, CDKN2A is directly inactivated through interactions with HPV viral proteins; the E7 viral protein inactivates Rb protein, which in turn upregulates expression of p16, thereby promoting cell cycle progression (see Fig. 73.3 ).
The mitogenic pathway is a broadly regulated pathway and the target of the first monoclonal antibody used to treat HNSCC, cetuximab. Its members include EGFR, Ras, phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), and phosphatase and tensin homolog (PTEN). Cetuximab, the EGFR-targeted monoclonal antibody, was the first FDA approved targeted therapeutic for HNSCC. Additional therapeutics targeting PI3K, mTOR, and additional downstream components of this pathway, are currently under active investigation.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here