Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
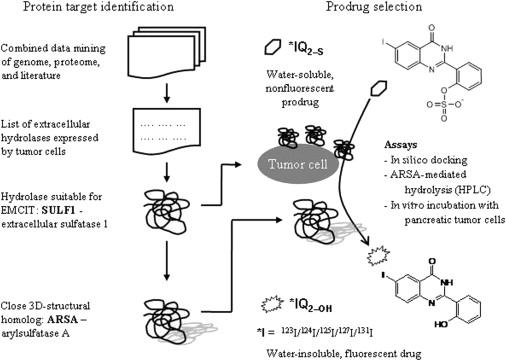
The concept of enzyme-mediated cancer imaging and therapy (EMCIT) involves the use of an enzyme specifically overexpressed on the surface of cancer cells. As such, the enzyme can act as a mediator for the hydrolysis of a soluble, radioisotopically labeled prodrug to a water-insoluble drug. This enzyme-dependent and site-specific hydrolysis provides a noninvasive technique for imaging and therapy, based on the rapid in vivo precipitation and entrapment of water-insoluble radioactive molecules within the extracellular spaces of solid tumors and minimal uptake into normal tissues.
The working prototype for the EMCIT approach was first developed and confirmed for alkaline phosphatase (ALP) , a hydrolase with monophosphoesteric activity that is known to be overexpressed on the plasma membranes of many tumor-cell types including breast and ovarian carcinoma . In 2008, the EMCIT concept was also successfully established for prostatic acid phosphatase (PAP) in prostate cancer . In both cases, the radiolabeled prodrug, a derivative of quinazolinone, was shown to have very EMCIT-suitable properties. Namely, ammonium 2-(2′-phosphoryloxyphenyl)-6-iodo-4-(3 H )-quinazolinone (IQ 2–P ) was shown to be hydrolyzed by ALP and PAP to the water-insoluble, fluorescent 2-(2′-hydroxyphenyl)-6-iodo-4-(3 H )-quinazolinone (IQ 2–OH ) . In vitro incubation of 125 IQ 2–P / 127 IQ 2–P with several human and mouse tumor cell lines (e.g., breast, colon, lung, ovary, and prostate) resulted in the efficient and rapid transformation of the prodrug to the corresponding water-insoluble derivative, which was effectively retained on the surface of cancer cells. The prodrug was minimally hydrolyzed in the presence of normal human mammary epithelial cells (HMEC) and mouse liver, spleen, kidney, and muscle cells . Importantly, IQ 2–P can be readily radiolabeled with an isotope having decay characteristics suitable for single-photon emission computed tomography (SPECT) or positron emission tomography (PET) imaging (e.g., 123 I, 124 I) or for therapy (e.g., 131 I).
Here we present the application of the EMCIT approach to pancreatic cancer. To a large extent, this chapter is based on the results of our previously published results . In the United States, it is estimated that 45,220 men and women will be diagnosed with pancreatic cancer in 2013, and an estimated 38,460 patients will die from this disease in 2013 ( www.cancer.gov/cancertopics/types/pancreatic ). Despite the lethal nature of pancreatic tumors, the possibility of early diagnosis is practically nonexistent. Thus, the development of technologies that enable noninvasive sensing of this disease and therapeutic intervention at an early stage is clearly desirable. Therefore, in analogy with our earlier investigations in which the overexpression of ALP was utilized to target such iodoquinazolinone derivatives to various tumors (e.g., colon, lung, ovarian) and PAP in prostate tumors, the intention of our studies is to (1) identify an enzyme that is overexpressed by pancreatic cells and that is able to hydrolyze the quinazolinone prodrug; (2) design, synthesize, and characterize an iodoquinazolinone analog that is a substrate for the extracellular pancreatic cancer–identified hydrolase and with EMCIT-suitable characteristics; and (3) perform in silico as well as in vitro binding assays of such derivatives with the data-mining-identified target prior to in vivo assessment.
In order to investigate a pancreatic target candidate, we have drawn on our recently developed bioinformatics methods of data mining that are based on the combined exploration of scientific literature, gene–protein databases, and knowledge-pathway databases . Using this approach, a new target that has been identified is an enzyme overexpressed in human pancreatic cancer called extracellular sulfatase 1 (SULF1). The primary activity of SULF1 is the desulfation of heparan-sulfate-proteoglycan (HSPG) . This sulfatase has also been shown to regulate the growth of pancreatic cancer cells and is overexpressed in pancreatic cancer . The enzyme belongs to the arylsulfatase family, whose members have the ability to desulfate molecules with sulfate-attached aromatic rings, e.g., para -nitrocatechol sulfate (pNCS) . Because of this activity, and in analogy with the arylphosphatase activity of ALP and PAP, we have modified IQ 2–P to its sulfur-derived analog 2-(2′-sulfooxyphenyl)-6-iodo-4-(3 H )-quinazolinone (IQ 2–S ), performed modeling studies, characterized the compound, and determined its hydrolysis in silico and in vitro.
This chapter is mainly based on our published paper . We briefly present how the SULF1 was identified using the combined data mining approaches. We then carried out the in silico molecular docking to predict the potential enzymatic selectivity for the analogs followed by in vitro incubation of the three enzymes (arylsulfatase A (ARSA), ALP, and PAP) with sulfate- and phosphate-quinazolinone derivatives. To further prove that the hydrolysis of these prodrugs occurs on the surface of the cancer cells, T3M4 pancreatic cancer cells (as well as other cancer cell lines: ovarian cancer cells OVCAR-3 and prostate cancer cells LNCaP) were incubated in vitro with the iodoquinazolinone derivatives and showed that their hydrolysis leads to the formation and precipitation of 127 IQ 2–OH fluorescent crystals on the cell surface. These findings were the first to report the targeting of a radioactive substrate to SULF1 and revealed that such structures would be useful in imaging ( 123 I/ 124 I/ 131 I) and radiotherapy ( 131 I) of pancreatic cancer.
The search for hydrolases expressed in the extracellular space of pancreatic cancer cells was completed using our data mining strategy. This approach in a combined manner explores scientific literature, gene and protein databases, and pathway knowledge bases. Our first strategy published in 2006 utilized the text mining software LSGraph and the pathway knowledge bases of Ingenuity Pathways Analysis ® (IPA) (version 2.0) . This combined approach of data mining identified 450 proteins related to pancreatic cancer from which one relevant target, SULF1, was identified (see following) .
In 2008, we published the data mining approach based on the knowledge base of IPA including the exploration of the cancer microarray platform Oncomine™ ( oncomine.org ), in order to identify extracellular hydrolases in cancer cells and blood-borne biomarkers for early diagnosis of cancer . Oncomine™ was chosen because it is a cancer microarray platform incorporating over 670 public independent microarray datasets totaling more than 73,000 samples, which span 20 major cancer types (September 2013). It unifies a large compendium of other published cancer microarray data (Gene Expression Omnibus (GEO; ncbi.nlm.nih.gov/geo)) and Stanford Microarray Database ( smd.princeton.edu ), and uniquely provides differential expression analyses comparing most major types of cancer with their respective normal tissues. For example, to identify potentially important genes in a particular cancer, users can perform a “cancer vs. normal” analysis for a given cancer type (e.g., pancreas) and those genes that are upregulated in cancer relative to its normal tissue can be retrieved as a list . Recently, we checked the presence of SULF1 in the oncomine.org database and found that in three out of seven microarray datasets, the gene of this enzyme meets the threshold of being in the top 1% genes of cancer vs. normal datasets. Thus, using this more recent approach, SULF1 would have been identified as the potential target as well.
Extracellular SULF1 is a potential EMCIT target ( Figure 17.1 ). While this enzyme is situated in the endoplasmic reticulum and Golgi stack, it has been shown, by its similarity to homologous proteins, to be secreted outside the cell (UniProt ID: SULF1_HUMAN; Entrez Gene ID: 23213). As indicated in protein and gene databases, its localization in the extracellular space infers from direct assays . Thus, the combined data mining approach used brings together the information on enzyme function, localization, ontology, and known expression in pancreatic cancer microarray datasets.
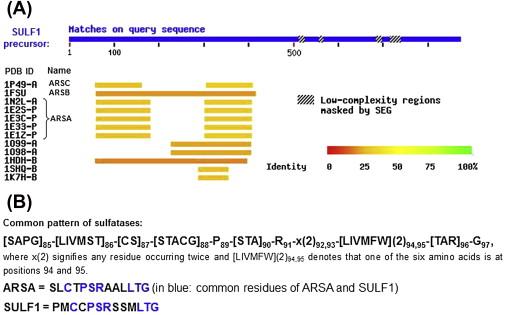
In order to situate the selected enzyme SULF1 with respect to protein family and to determine the closest available structural homologs, sequence alignments against known protein sequences of the UniProt Knowledgebase 9.2 ( uniprot.org ) were queried using BLAST network service of the Swiss NCBI BLAST program reference (PMID:9254694) . The blastp option was set as restricted to mammalian taxon, searched only in UniProt-curated sequences and excluding fragments. The assessment of SULF1 against sequences of proteins with experimentally determined 3-D structures was performed against nonredundant sequences of the Protein Data Bank (PDB, rcsb.org ) also using NCBI BLAST2 software ( Figure 17.2 ). Furthermore, sequence alignment of SULF1 against all mammalian proteins in the UniProt database placed SULF1 into the “alkaline-phosphatase-like clan” (Interpro ID: IPRO17849; ebi.ac.uk/interpro/entry/IPR017849). Such findings encouraged us once more to consider SULF1 for the EMCIT concept as this approach was demonstrated to work for the prostatic acid and ALPs .
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here