Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The pathophysiology of atrial fibrillation (AF) remains incompletely characterized; however, epidemiologic studies demonstrate a heritable basis for the arrhythmia. In recent years, the appreciation of AF heritability has stimulated the search for the genetic underpinnings of the disease. Genetic mapping studies have identified rare mutations and common variants associated with AF. In addition to validating suspected electrophysiologic mechanisms underlying AF, recent genetic discoveries have identified previously unrecognized pathways involved in the development of AF. Investigators are now searching for causal variants at many identified loci, investigating the biologic mechanisms linking AF susceptibility loci to disease and assessing the clinical implications of recent genetic discoveries.
Reports of familial clustering of AF date back to the early 1940s. , Familial AF was generally regarded as a rare condition for many years thereafter. However, over the past decade a major paradigm shift occurred with the widespread recognition of the heritability underlying AF. In the community-based Framingham Heart Study, 27% of individuals with AF had a first-degree relative with AF confirmed by electrocardiography. Familial AF was associated with a 40% increased risk of AF to another family member over a subsequent 8-year period ( Fig. 47.1 ). The risk associated with familial AF remained even after adjusting for established clinical risk factors for AF. A study from Denmark demonstrated that AF was more common among monozygotic twins compared with dizygotic twins, implicating a genetic predisposition for the arrhythmia even among those raised with shared environmental exposures.
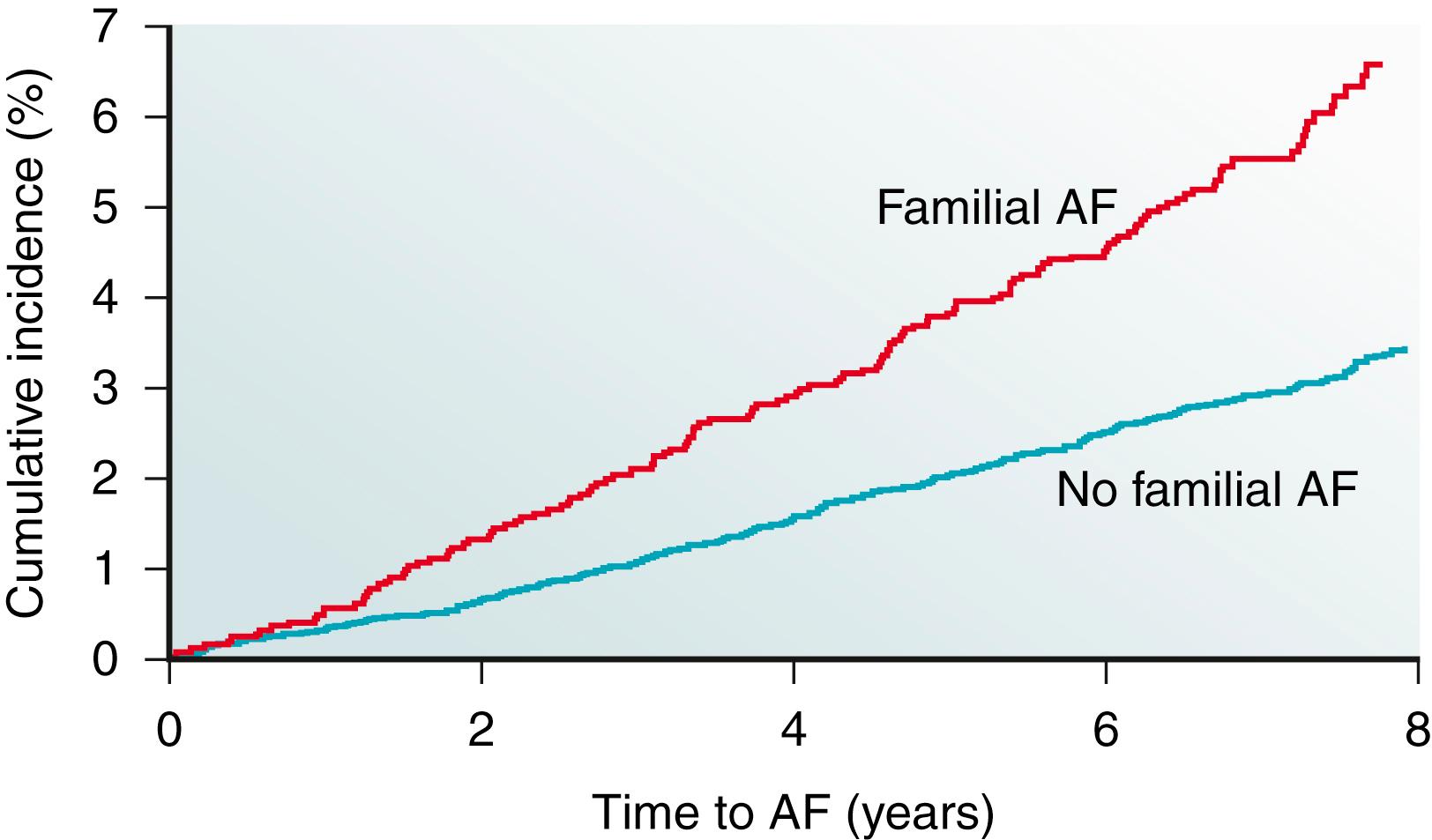
In numerous reports, the heritability of AF appears to be greatest among younger individuals , , and those without structural heart disease. , Premature familial AF, or that occurring in family members age 65 years or younger, was associated with a twofold increase in the risk of AF compared with individuals without familial AF in the Framingham Heart Study. Nevertheless, data from Framingham provide evidence that the heritability of AF is present in the elderly as well. More recent approaches using a genetic approach determined that the heritability of AF is approximately 22%.
Early attempts to identify the genetic basis of AF used classic linkage analysis to identify monogenic disease loci in families. Linkage analysis relies on the fact that genetic recombination during meiosis shuffles regions of the genome. Assessing genetic markers at known locations of the genome can help determine which loci transmit along with disease.
The first genetic locus for AF was described in 1997 by Brugada and associates who identified an AF susceptibility region on chromosome 10 in a large family with autosomal dominant AF ( Table 47.1 ). Since this initial report, linkage analysis has occasionally been used to identify genetic mutations underlying AF in a number of large families with AF.
| Gene | Gene Product | Presumed Mechanism of Action | References |
|---|---|---|---|
| Potassium Channels | |||
| KCNH2 | α-Subunit of K V 11.1 channel (HERG) | Modulation of I Kr | 68, 69 |
| KCNQ1 | α-Subunit of K V 7.1 channel | Increased I Ks | 10, 12, 70–75 |
| KCNE1 | β-subunit of K V 7.1 channel (minK) | Increased I Ks | 76, 77 |
| KCNE2 | β-Subunit of K V 7.1 channel (MiRP1) | Increased I Ks | 78, 79 |
| KCNE5 | β-Subunit of K V 7.1 channel | Increased I Ks | 80 |
| KCNJ2 | β-Subunit of K ir 2.1 channel | Increased I K1 | 81–83 |
| KCNJ3 | α-Subunit of K ir 3.1 channel | Increased I KACh | 18 |
| KCNA5 | α-Subunit of K V1.5 channel | Modulation of I Kur | 78, 84, 85 |
| KCNJ5 | α-Subunit of K ir 3.4 channel | Increased I KACh | 18 |
| KCNJ8 | α-Subunit of K ir 6.1 channel | Increased I KATP | 86 |
| KCND3 | α-Subunit of K V4.3 channel | Increased I to current | 87, 88 |
| ABCC9 | SUR2A subunit of K ATP channel | Decreased I KATP | 89 |
| KCNN3 | Ca-activated K channel KCa2.3 | Modulation of SK Ca current | 90, 91 |
| Sodium Channels | |||
| SCN5A | α-Subunit of Na V 1.5 | Modulation of I Na | 92–98 |
| SCN10A | α-Subunit of Na V 1.5 | Potential modulation of Na v 1.5 | 99, 100 |
| SCN1B | β-Subunit of Na V 1.5 | Reduced I Na | 101, 102 |
| SCN2B | β-Subunit of Na V 1.5 | Reduced I Na | 101 |
| SCN3B | β-Subunit of Na V 1.5 | Reduced I Na | 92, 103 |
| SCN4B | β-Subunit of Na V 1.5 | Not characterized | 104, 105 |
| Other Ion Channels or Related Proteins | |||
| HCN4 | Hyperpolarization activated cyclic nucleotide gated K channel 4 | Modulation of the pacemaking I f current | 106, 107 |
| CAV1 | Caveolin 1 | Altered ion channel signaling | 108, 109 |
| CLCN2 | Chloride channel protein 2 | Loss of channel function | 110 |
| KCND2 | α-Subunit of K V 4.2 | Enhanced I to current | 111 |
| Gap Junction Proteins | |||
| GJA1 | Connexin43 | Reduced intercellular electrical coupling | 112 |
| GJA5 | Connexin40 | Impaired intercellular electrical coupling | 113–115 |
| Transcription Factors | |||
| GATA4 | GATA4 | Decreased activity | 116–118 |
| GATA5 | GATA5 | Decreased activity | 119–121 |
| GATA6 | GATA6 | Decreased activity | 122–124 |
| NKX2-5 | NKD2.5 | Decreased activity | 125–127 |
| PITX2 | PITX2 | Decreased activity | 128–131 |
| SHOX2 | SHOX2 | Decreased activity | 132–134 |
| ZFHX3 | ATBF-1 | Altered binding activity | 128 |
| Sarcomeric Genes | |||
| MYH6 | Myosin heavy chain 6 | Loss of function | 62, 135 |
| MYL4 | Myosin light chain 4 | Loss of function | 136–140 |
| TTN | Titin | Loss of function | 63–65 |
| Other Genes | |||
| C9orf3 | Aminopeptidase-O | Unclear | 128, 141 |
| JPH2 | Junctophilin-2: modulation of RyR activity | Decreased RyR stabilization | 142 |
| NPPA | Natriuretic peptide precursor A | Altered ANP signaling | 17, 143, 144 |
| NUP155 | Nucleoporin | Reduced nuclear membrane permeability | 145 |
| PLEC | Plectin | Reduced function | 61 |
| RYR2 | Ryanodine receptor | Increased activity | 146, 147 |
| SYNE2 | Neseprin-2 | Unclear | 128, 141 |
| SH3PXD2 | Tks5 | Enhanced macrophage migration | 148 |
| THRB | Thyroid hormone receptor β | Thyroid hormone resistance | 149–153 |
Chen and colleagues narrowed an AF susceptibility locus to a region on chromosome 11p15 in a family with autosomal dominant AF spanning four generations. The investigators extended their analysis by sequencing KCNQ1, a candidate gene in the region that encodes a potassium channel α-subunit. In the first transmembrane segment of the channel they discovered a highly conserved serine residue that was mutated to a glycine in all affected family members. Subsequent characterization revealed that the mutant protein increases the repolarizing I Ks current density when expressed with the KCNE1 subunit. This gain-of-function mutation is expected to result in shortened atrial refractory periods, thereby promoting reentry, a well-founded mechanism underlying AF. Our group led by Das and coworkers used a similar approach to map a mutation in the third transmembrane domain of KCNQ1. The S209P variant demonstrated a similar gain-of-function effect again implicating KCNQ1 as a disease susceptibility gene for AF.
Linkage analysis also has identified mutations in SCN5A that segregate with AF, conduction abnormalities, cardiomyopathy, and possibly early-onset ischemic stroke. SCN5A encodes a sodium channel α-subunit responsible for the depolarizing I Na current. The D1275N mutation that results in an aspartic acid for asparagine substitution was independently identified by different investigators, and associates with atrial standstill when cosegregating with connexin40 ( GJA5 ) mutations.
In another large multigenerational family with AF, Hodgson-Zingman and colleagues mapped a frameshift mutation to NPPA, which encodes atrial natriuretic peptide. The 2-bp deletion eliminates a stop codon and results in an additional 12 amino acids at the carboxy terminus of the mature 28-residue long ANP protein. Ex vivo rabbit hearts bathed in the mutant peptide demonstrated significantly shortened atrial effective refractory periods compared with those bathed with a wild-type atrial natriuretic peptide, again consistent with reentry as a pathogenic model for AF.
In aggregate, linkage analysis has implicated potassium ( KCNQ1 , and KCNJ3 ) and sodium ( SCN5A ) channel mutations, a nuclear envelope protein ( NUP155 , ), NPPA, and loci on chromosomes 6 and 10. , However, the large multigenerational families needed for linkage analysis remain rare. More often AF is observed in smaller families; thus is it is often hard to establish the causality of apparently disease-causing mutations with such limited genetic information.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here