Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The human gastrointestinal (GI) tract represents one of the largest interfaces between the host, environmental factors, and antigens. In addition to food and liquids, an abundance of environmental microorganisms and xenobiotics pass through the GI tract daily, posing a potential threat on intestinal health. The collection of bacteria, fungi, viruses, and yeasts in the GI tract, termed gut microbiota, can be considered a microbial organ placed inside the GI tract. The gut microbiome, the gut microbiota along with genomic material, has coevolved with humans over thousands of years to form a mutualistically beneficial relationship. Through a range of physiologic and metabolic functions, the gut microbiota offers many benefits to the host such as maintaining gut integrity, harvesting energy, providing protection against pathogens, and regulating host immunity.
The human body is colonized with microbes in numbers that are equal to, if not greater than, our somatic cells. It is estimated that 500 to 1000 bacterial species exist in the human body at any one time, and the number of subspecies is far greater. Each strain of bacteria has its own genome composed of thousands of genes, thus with an estimated 1000 gut bacterial species and 2000 genes per species, that is 100 times more genetic diversity than the human genome, which is estimated at 20,000 genes. Combined data from the MetaHit and the Human Microbiome Project have provided the most comprehensive view of the human-associated microbial repertoire to date. Data from these projects, in which 2172 isolated species were classified into 12 different phyla, indicated 93.5% belonged to Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. In humans, 386 strictly anaerobic species were identified to be localized around mucosal regions of the oral cavity and GI tract. We are learning that each individual has a unique microbiome, and there are great differences between individuals. , However, it should also be noted that microbiota that differ in terms of composition may share some degree of functional redundancy, yielding similar protein or metabolite profiles. This information is crucial for developing therapeutic strategies to modify and shape the microbial community in disease. How overall wellness or disease progression is influenced by these variations, within or between people over time, is not fully understood. What we do know is that many illnesses are associated with shifts in the microbiome and its metabolome, which is correlated with altered communication with the host immune, endocrine, and nervous system functions.
Molecular identification using next-generation high-throughput sequencing technologies has replaced traditional culturing, therefore allowing a window into the diversity of microbial communities. Ribosomal RNA (rRNA) gene sequence analysis is a powerful method for identifying and quantifying members of microbial communities. Present in all living organisms and containing diverse species-specific domains, this gene contains nine highly variable regions (V1 to V9), which allows species to be easily distinguished, and thus can be used to reliably assume phylogenetic relationships at multiple taxonomic levels. Most studies are currently based on sequencing of the 16S rRNA gene, which has several highly conserved regions interleaved with variable regions. Conserved regions serve as “anchors” for designing polymerase chain reaction (PCR) primers, while sequencing the variable regions identifies the bacteria. Amplification, sequencing, and comparison to databases allow for the identification of bacterial lineages and their proportions in a community. The new deep sequencing methods facilitate more efficient and inexpensive data acquisition, but optimal methods are less well developed.
Both fecal samples and mucosal biopsies can be used for analysis of the human gut microbiota to quantify the bacterial taxa present. Feces have been shown to contain a representative collection of the bacterial taxa from the lower GI tract, permitting convenient sampling for characterization of the gut microbiota. Linking the human microbiome to GI disease often requires large sample sizes, so there is a need for practical specimen acquisition methods that allow analysis of large numbers of human subjects, focusing attention on methods for collecting and analyzing fecal samples
During human microbiome development, each body site develops a specific biogeography. The skin, which is covered with microbes, has site-specific microbiome composition and structure. The physical and topographical characteristics of skin play a role in shaping the individuality of the microbiome so that each person develops a unique microbial signature on their skin irrespective of the differences between skin sites. Regarding degree of stability, the vaginal microbiome is similar to the skin microbiome, and it has been shown to cluster with diseases. In asymptomatic women, the vaginal microbiota is more likely dominated by individual species of Lactobacillus and additional diverse anaerobic taxa. The lactic-acid bacteria may then benefit the host by maintaining a lower vaginal pH and thus prevent the likelihood of pathogen colonization. Microbial variation within an individual woman does occur over days to weeks, although menstruation and pregnancy appear to result in a similar microbiome in different groups of women. Prior belief that the lungs are sterile has been refuted. Next-generation sequencing has identified a distinct microbiome in healthy lungs, characterized by the prevalence of bacteria from the phylum Bacteroidetes .
Traditionally the intestinal tract has been considered sterile at birth; however, emerging data suggest that colonization begins in utero, because the presence of microorganisms has been identified in amniotic fluid, fetal membranes, umbilical cords, placentas, and meconium. Moles et al. characterized the microbial composition of spontaneously released meconium and feces of preterm infants, born at a gestational age of 32 weeks or earlier and/or with a birth weight 1200 g or less, and without malformations or metabolic diseases, during their first month of life. Although both samples had positive bacterial cultures demonstrating interindividual differences, meconium was distinct from fecal samples. Bacilli and other Firmicutes were the main bacteria groups detected in meconium, and Proteobacteria dominated in the fecal samples. Another study that investigated samples of umbilical cord blood collected from 20 healthy neonates born by cesarean section (C-section) identified isolates belonging to the genus Enterococcus, Streptococcus, Staphylococcus, or Propionibacterium . Although further study is needed, there is increasing evidence that microorganisms inhabit the placenta, amniotic fluid, and umbilical cord; thus, by swallowing amniotic fluid in utero, the fetus begins to colonize the developing GI tract.
Colonization continues with the birthing process, with the gut microbiota of newborns closely resembling the microbes encountered during birth. Using 16S rRNA gene pyrosequencing, an analysis was performed of newborn term babies to characterize bacterial communities from mothers and their newborn babies to gain community-wide perspective on the influence of delivery mode and body habitat on the infants’ first microbiota. Samples from mothers’ skin, oral mucosa, and vagina were obtained 1 hour prior to delivery, and infants’ skin, oral mucosa, and nasopharyngeal aspirate were sampled less than 5 minutes after birth and meconium less than 24 hours after delivery. Interestingly, as opposed to the mothers’ highly differentiated communities, regardless of delivery mode the infants’ bacterial communities were undifferentiated across multiple body sites. Vaginally delivered infants acquired bacterial communities resembling their own mother’s vaginal microbiota, which was dominated by Lactobacillus , Prevotella , or Sneathia spp. In contrast to vaginal delivery, babies birthed by C-section harbored bacterial communities that resembled those of the skin, comprising Staphylococcus , Corynebacterium , and Propionibacterium spp. , Analysis of these communities found that they were similar to other women’s skin flora, indicating that the baby is in contact with other humans. Because C-section babies do not receive that first vaginal maternal inoculum, it not only affects the development of the GI microbiota, , but it may also explain the increased vulnerability C-section–delivered infants have to certain pathogens. Compared with vaginally delivered babies, C-section–delivered babies have a higher incidence of atopic diseases, allergies, asthma, , and skin infection with methicillin-resistant Staphylococcus aureus (64% to 82% of C-section babies).
The first major phase of gut colonization begins with infant feeding. Although the newborn’s gut microbiota is composed of relatively few species and lineages, diversity rapidly increases within the first few years of life. , Exactly why there is an increase in diversity is unknown, but it is hypothesized to be from an enlarging intestinal tract that provides a larger habitat for more bacteria, increased exposure to environmental bacteria, and/or dietary changes.
Meeting all nutritional and physiologic requirements, breast milk is the optimal food for infants. Human milk contains protein, fat, and carbohydrate, as well as immunoglobulins and endocannabinoids. Breastfeeding has been shown to enrich vaginally acquired, lactic acid–producing bacteria in the baby’s intestine. Breast milk is not sterile, because it contains as many as 600 different species of bacteria, including beneficial Bifidobacterium species B. breve, B. adolescentis, B. longum, B. bifidum, B. catenulatum, and B. dentium . All of these Bifidobacterium species may promote healthy microbiota development.
The carbohydrate component of human milk, in addition to lactose, also contains oligosaccharides, which compose the third largest solid component. Human milk oligosaccharides are indigestible polysaccharides, formed by a small number of different monosaccharides. These oligosaccharides serve as prebiotics by selectively stimulating growth of members of the genus Bifidobacterium . An increased proportion of Bifidobacteria is noted in breast-fed infants compared with those fed formula. Bifidobacteria have been linked with strengthening gut mucosal protection through activities against pathogens. Correlated with modulation of the intestinal immune system , production of immunoglobulin A has been shown to be induced by Bifidobacteria .
Schwartz et al. studied breast-fed and formula-fed infants and their mothers. Stool samples from each infant were collected, and microbial DNA was extracted and sequenced; mRNA was isolated from stool containing host gut–exfoliated epithelial cells and was processed for microarray analysis. Results indicated that the microbiome of breast-fed infants was significantly enriched in genes associated with virulence functionality. In addition, a multivariate correlation between the gut microbial genes associated with bacterial pathogenicity and the expression of host genes associated with immune and defense mechanisms was made. Interestingly, the operational taxonomic unit (OTU) composition and genetic potential of the microbiota differed between breast-fed and formula-fed infants. The researchers suggested their findings indicate human milk promotes the mutualistic crosstalk between the mucosal immune system and the microbiome in the maintenance of intestinal homeostasis. More research is needed to better determine the mechanisms by which Bifidobacteria produce these effects.
Aerotolerance of the intestinal microbiota also seems to differ between breast-fed and formula-fed infants. Aerobic organisms are more common in the feces of breast-fed infants, whereas anaerobic and facultatively anaerobic organisms, which preferentially use anaerobic glycolysis, are more frequently identified in feces of formula-fed infants. Bacteroides and Clostridia colonization differs between the two types of feeding, with breast-fed infants characterized by lower concentrations of both.
With a better understanding of the composition of human breast milk, developments of complex infant formulas have attempted to mimic its nutritional value with the goal of creating an acceptable alternative for mothers unable to breast-feed. Infant formulas are not a perfect substitute for human milk because bioactive compounds contained in breast milk known to affect nutrient absorption and digestion, immune protection, and defense against potentially pathogenic microbes are lacking. Unfortunately it is difficult to mimic the actions of these bioactive compounds. Although infants fed a formula enriched with oligosaccharides have been shown to harbor more Bifidobacteria in the feces, more evidence is needed for affirmation that infant formulas designed to mimic breast milk are beneficial.
The second transition in gut colonization occurs when the diet transitions to solid foods. Advancing the diet from simple solid foods (e.g., rice cereal) to one including more complex plant polysaccharides (e.g., peas) changes the relative ratio of bacterial phyla in the GI tract from an unstable community dominated by Actinobacteria and Proteobacteria to a stable mixture dominated by Firmicutes and Bacteroidetes , similar to that of an adult-like microbiota. Analysis of the gut microbiota from African and Italian children found that African children had an increased abundance and diversity of Bacteroidetes . This raises the possibility that the gut microbiota is adapted by local diets and that the gut microbiota can adapt via genomic changes to alterations in the host’s diet; this adaption can be achieved by incorporating genes from bacteria in the environment. Thus, following weaning and transitioning to solid foods, shaping of the gut microbiota moves towards that which is characteristic of an adult by three years of age.
The gut microbiota differs in numbers and composition along the length of the GI tract, reflective of the physiologic properties in a given region. Chemical, nutritional, and immunologic gradients along the gut affect the diversity and density of the gut microbiota. Although the saliva may contain up to 10 9 microbial cells/mL, low gastric pH limits bacterial numbers in the stomach and discourages ingested bacteria from entering the small intestine. In addition, due to bile and peristalsis, the jejunum is typically colonized only by relatively simple, rapidly growing, facultative anaerobes with the ability to adhere to epithelia/mucus. In the ileum, microbial numbers and diversity increase as peristalsis and acidity decrease. The colon is where the numbers and diversity are greatest, due to slow transit time and ample food supply from nutrients that have escaped digestion and absorption in the proximal intestinal tract. Overall microbiota vary between individuals, but up to 10 11 to 10 14 primarily anaerobic bacterial cells/g of feces are commonly detected in the colon. Prevotellaceae, Lachnospiraceae, and Rikenellaceae have been shown to dominate in the colon ( Fig. 4.1 ).
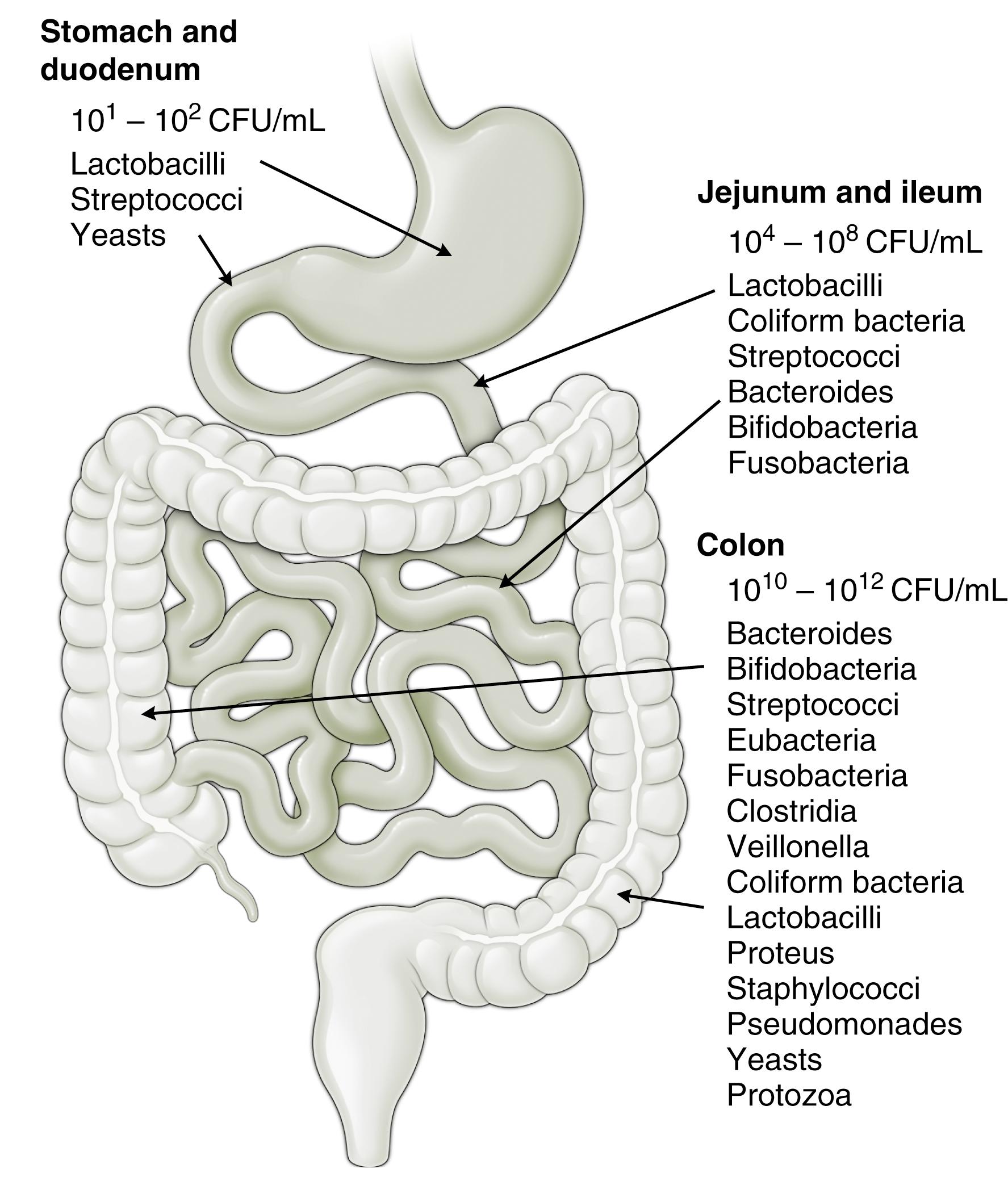
Although marked interpersonal microbial variability exists, a conserved spatial structure to the colonic microbiota, differentiating the luminal and mucosal communities, is noted even during periods of localized inflammation. On the other hand, fecal/luminal and mucosal compositions are significantly different, with Bacteroidetes abundance higher in fecal/luminal samples than in the mucosa. , Interestingly, mouse experiments show that bacterial species present in the outer mucus layer differ in proliferation and resource utilization compared with the same species in the intestinal lumen. This becomes important when considering sampling methods and locations for microbiota composition identification.
Despite prior thought surrounding an abundance of key organisms being present in all individuals, it appears there is more similarity among microbial genes, defining the microbiome more on a functional rather than organism level.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here