Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Collect specimens from site of infection before initiating therapy.
Collect an adequate volume of sample for testing required.
Tissue, fluid, or aspirates are always superior to a swab specimen. The only exception is collecting culture material from a hard-to-reach spot such as a throat or cervix.
Use required collection and transport materials to preserve specimen integrity.
Communicate clear orders and source information.
Expedite the transport of specimens to the laboratory and do not allow them to sit in collection areas.
We gratefully acknowledge the contributions of Ann C. Croft and Gail L. Woods, who authored this chapter in previous editions.
Appropriate specimen collection, transport, and processing are crucial preanalytical steps in the accurate diagnosis of infectious diseases. Guidelines for specimen handling are discussed in this chapter. General principles are reviewed first followed by discussion of the most common types of specimens submitted to the clinical microbiology laboratory for testing.
For optimal detection of the pathogens responsible for an infectious disease, specimens should be collected at a time when the likelihood of recovering the suspected agent is greatest. For example, the likelihood of recovering most viruses is greatest in the acute phase of the illness. Specimens for recovery of bacteria should ideally be collected before antimicrobial therapy is started.
The volume of specimen collected must be adequate for performance of the microbiological studies requested. If insufficient volume is received, the health care worker caring for the patient should be notified; either an additional sample can be obtained, or the physician must prioritize the requests.
Very little specimen is obtained with a swab, and much of the specimen is retained within the swab tip. Swabs should not be used as collection devices unless the specimen source is the throat, cervix, or other difficult-to-reach area. If a swab is used to collect the specimen, a polyester-tipped swab on a plastic shaft is acceptable for most organisms. Calcium alginate should be avoided for collection of samples for viral culture because it could inactivate herpes simplex virus (HSV), cotton may be toxic to Neisseria gonorrhoeae , and wooden shafts should be avoided because the wood may be toxic to Chlamydia trachomatis . Swabs are not optimal for detection of anaerobes, mycobacteria, or fungi, and they should not be used when these organisms are suspected. An actual tissue sample or fluid aspirate is always superior to a swab specimen for the recovery of pathogenic organisms.
Specimens should be obtained from the site of infection with minimal contamination from adjacent tissues and organ secretions, and with the exception of stool, should be collected in a sterile container. All specimens should be labeled with the name and identification number of the person from whom the specimen was collected, the source of the specimen, and the date and time it was collected.
After collection, specimens should be placed in a biohazard bag and transported to the laboratory as soon as possible. If a delay is unavoidable, urine, sputum and other respiratory specimens, stool, and specimens for detection of C. trachomatis or viruses should be refrigerated to prevent overgrowth of normal flora. Cerebrospinal fluid (CSF) and other body fluids, blood, and specimens collected for recovery of N. gonorrhoeae should be held at room temperature because refrigeration adversely affects recovery of potential pathogens from these sources.
Each laboratory director must establish criteria for rejecting specimens unsuitable for culture. Most clinical microbiologists agree that the following specimens should be rejected:
Any specimen received in formalin
24-hour sputum collections
Specimens in containers from which the sample has leaked
Specimens that have been inoculated onto agar plates that have dried out or are outdated
Specimens contaminated with barium, chemical dyes, or oily chemicals
Foley catheter tips
Duplicate specimens (except blood cultures) received in a 24-hour period
Blood catheter tips submitted for patients without concomitant positive blood culture
The following specimens should be rejected for anaerobic culture:
Gastric washings
Urine other than suprapubic aspirate
Stool (except for recovery of Clostridium difficile for epidemiologic studies or for diagnosis of bacteria associated with food poisoning)
Oropharyngeal specimens except deep tissue samples obtained during a surgical procedure
Sputum
Swabs of ileostomy or colostomy sites
Superficial skin specimens
Safety is the responsibility of the laboratory director and, per the Clinical Laboratory Improvement Amendments (CLIA), cannot be delegated to others. The laboratory director should collaborate with infection control, institutional safety committees, environmental services, engineering, and others within the hospital and department to ensure that there are current and necessary policies and procedures, engineering controls, personal protective equipment (PPE), and a trained work force. The laboratory director is also responsible for making sure the policies and procedures are followed. Specimen processing can have additional challenges because this workforce often has the least amount of formal technical education.
Universal Precautions were designed to protect workers from infectious substances in blood and body fluids. Body Substance Isolation Precautions were designed to protect workers from transmission of organisms from moist body surfaces. In 1996, the US Hospital Infection Control Practices Advisory Committee unified these into “Standard Precautions.” As described in Biosafety in Microbiological and Biomedical Laboratories (BMBL), Standard Precautions apply to (1) blood; (2) all body fluids, secretions, and excretions except sweat regardless of whether or not they contain visible blood; (3) nonintact skin; and (4) mucous membranes ( ). Standard Precautions must be followed when handling all specimens. A risk assessment should be performed in each laboratory area; it should detail the particular risks with each procedure. The safety recommendations for engineering controls, PPE, and work practices can be tailored to the risk ( ). Appropriate barriers are used to prevent exposure of skin and mucous membranes to the specimen. Gloves and a lab coat must be worn at all times when handling patient specimens, and masks, goggles (or working behind a plastic shield), and impermeable gowns or aprons must be worn when there is a risk for splashes or droplet formation. Optimally, all specimen containers, but at a minimum, those containing respiratory secretions and those submitted specifically for detection of mycobacteria or fungi should be opened in a biological safety cabinet. Specimens collected for virus isolation should be handled in a biological safety cabinet to prevent contamination of the cell cultures.
When specimens or cultures must be shipped to a reference laboratory, they must be packaged according to dangerous goods shipping guidelines (see International Air Transport Association website, available at http://www.iata.org/whatwedo/cargo/dgr/Pages/index.aspx ). Specimens must be limited to no more than 40 mL. Cultures of bacteria and fungi should be grown on solid media in tubes. The cap of the primary container (tube or vial) should be sealed with waterproof tape and inserted into a second container surrounded by sufficient packing material to absorb the entire volume of the culture or specimen if the primary container were to leak or break. If several primary tubes are placed in a second container, they must be either individually wrapped or separated so as to prevent contact between them, and there must be secondary packaging, which must be leakproof. The second container should be capped and placed in a shipping container made of corrugated fiberboard or hard plastic. An itemized list of contents must be enclosed between the secondary and outer packaging. The secondary and outer containers should be of sufficient strength to maintain their integrity at temperature and air pressures to which they will be subjected. If a specimen must be shipped on dry ice (which is considered to be a hazardous material), it must be marked “Dry ice, frozen medical specimen.” The dry ice should be placed outside the second container with the packing material in such a way that the container does not become loose inside the outer container as the dry ice evaporates. All infectious shipping packages must be labeled with an official label containing the address and contents as well as the name and telephone number of the person responsible for the shipment. All laboratorians who package and ship materials that are known or reasonably expected to contain a pathogen must have documentation of training. There are several commercial and government-based resources for training.
The rapid identification and susceptibility testing of bloodborne pathogens is one of the most critical functions of the microbiology laboratory. Even with the increased use of molecular technologies capable of identifying organisms and some markers of resistance from either blood specimens or positive blood culture bottles, blood cultures are still the “gold standard” for identifying bacteria responsible for bacteremia, sepsis, infections of native and prosthetic valves, suppurative thrombophlebitis, mycotic aneurysms, and infections of vascular grafts. Blood cultures also are useful in diagnosing invasive or disseminated infections caused by certain fungi, especially Cryptococcus neoformans , Candida spp., Fusarium spp., and Histoplasma capsulatum . Serologic testing or nucleic acid amplification tests (NAATs) are needed to identify two important bacterial causes of culture-negative endocarditis, Coxiella burnetii and Bartonella spp. Parasites are usually detected in blood by microscopic examination of peripheral smears, but enzyme immunoassay (EIA) and NAATs are also used.
Timely detection and accurate identification of organisms in the blood depend on appropriate collection, transport, and processing of the specimen. In general, two to three sets of blood cultures should be collected for identification and susceptibility testing before antimicrobial therapy is initiated. Consensus guidelines recommend peripheral venipuncture rather than draws from intravascular catheters. Good skin cleaning and phlebotomy technique minimize the presence of skin contaminants in blood cultures ( ). The use of iodine tincture, or chlorhexidine gluconate, is preferable to povidone–iodine solutions because the former require only about 30 seconds after application to achieve antiseptic effect; the latter requires up to 2 minutes. Each set of peripheral blood cultures must be drawn from a separate venipuncture site so that if a typical contaminating organism is present in one set and not the other, the organism can be more easily classified as a contaminant.
Compared with venipuncture, the risk for contamination is increased when blood cultures are drawn from an indwelling vascular device. Although it might seem advantageous to save the patient the discomfort of a venipuncture, a contaminated blood culture can cause even more diagnostic cultures, unnecessary antibiotics, and a prolonged stay in the hospital. If it is necessary to draw a culture through an intravenous line, another blood culture should be collected simultaneously from a venipuncture to aid in the interpretation of a contaminated line culture ( ).
The optimal time to draw blood for cultures when bacteremia or fungemia is suspected is just before a fever or chill, but this is not predictable. The single most important factor to detect bacteremia is volume of blood collected. In adults with bacteremia, the number of colony-forming units (CFUs) per milliliter of blood is frequently low. Therefore, for adults, collecting multiple sets of blood cultures with each bottle filled to optimal volume is most important. ( ). In infants and children, the concentration of microorganisms in blood is higher, and collection of 1 to 5 mL of blood per culture is adequate. When blood cultures are drawn by personnel other than phlebotomists, it can be a challenge to obtain sufficient volumes in the culture bottles. With the current focus on detecting and treating sepsis, it behooves the pathologist to collaborate with clinical colleagues to optimize the amount of blood collected and improve the ability of the blood culture to detect bacteremia ( ; ).
Recommendations concerning the number of blood specimens to collect are based on the nature of the bacteremia: transient, intermittent, or continuous . Transient bacteremia follows manipulation of a focus of infection (e.g., an abscess, a furuncle, or cellulitis), instrumentation of a contaminated mucosal surface (as occurs during dental procedures, cystoscopy, urethral catheterization, suction abortion, or sigmoidoscopy), or a surgical procedure in a contaminated site (e.g., transurethral resection of the prostate, vaginal hysterectomy, colon resection, and debridement of infected burns). Transient bacteremia also occurs early in the course of many systemic and localized infections such as meningitis, pneumonia, pyogenic arthritis, and osteomyelitis. Most intermittent bacteremias are associated with an undrained abscess, whereas continuous bacteremia is the hallmark of intravascular infection, such as bacterial endocarditis, mycotic aneurysm, or an infected intravascular catheter. Continuous bacteremia also occurs during the first few weeks of typhoid fever and brucellosis.
The optimal number of blood cultures for detection of bacteremia in patients without endocarditis is controversial. Most authorities agree that two or three 20-mL blood samples drawn over a 24-hour period and equally distributed into aerobic and anaerobic blood culture bottles will detect most bloodstream infections. One investigator demonstrated that 80% of bacteremias were detected with two blood cultures and 96% with three blood cultures. All bacteremias were detected with four blood cultures, but the routine collection of four blood cultures (up to 80 mL of blood) should be weighed against the risk for anemia ( ). The optimal time interval between cultures is unknown, but 30 to 60 minutes for the first two sets has been suggested, with another one to two sets drawn over the remaining 24 hours if symptoms of septicemia persist ( ). However, if initiation of antimicrobial therapy is deemed urgent, cultures should be collected before therapy is begun, from separate sites, within a few minutes.
Organisms such as the coagulase-negative staphylococci, viridans streptococci, Corynebacterium spp., Bacillus spp., and Propionibacterium spp. are frequent blood culture contaminants but may also be true pathogens. Collecting two sets of blood cultures per febrile episode helps distinguish probable pathogens from contaminants. If two sets are drawn from different venipuncture sites, the odds of both sets being contaminated by skin flora are very low. If two sets are drawn at the same time and only one set contains a skin contaminant, it is safe to assume that the culture was contaminated during collection. If only one set is drawn and a contaminant is present, it can be difficult to not treat the organism, especially if the patient has had recent surgery.
Host factors such as antibodies, complement, phagocytic white blood cells, and antimicrobial agents may impede recovery of microorganisms from blood; therefore, various approaches have been used to counteract these factors. Diluting the blood specimen in broth medium in a 1:10 ratio provides optimal neutralization of the serum bactericidal activity ( ). Incorporating 0.02% to 0.05% sodium polyanethol sulfonate in the blood culture medium inhibits coagulation, phagocytosis, and complement activation and inactivates aminoglycosides. Methods that counteract the presence of antimicrobial agents include using antibiotic-adsorbent resins or the lysis-centrifugation system.
Three commercially available automated blood culture systems, each with advantages and disadvantages, are available. These continuously monitored automated detection systems have essentially replaced manual systems. All systems use nutritionally enriched liquid media, which are capable of supporting growth of most bacteria and many pathogenic yeasts. Traditionally, two bottles, an aerobic and an anaerobic, are inoculated.
Three major automated continuously monitoring blood culture systems are available commercially in the United States. With these systems, the usual incubation period is 5 days ( ). The BacT/ALERT 3D system (bioMerieux) is based on the colorimetric detection of carbon dioxide (CO 2 ) produced during microbial growth. A CO 2 sensor is bonded to the bottom of each blood culture bottle and is separated from the broth medium by a membrane that is impermeable to most ions and to components of media and blood but freely permeable to CO 2 . Inoculated bottles are placed in cells in the instrument, which provides continuous rocking of both aerobic and anaerobic bottles. If bacteria are present, they generate CO 2 , which is released into the broth medium; the pH then decreases, causing the sensor to change color from green to yellow. Color changes are monitored once every 10 minutes by a colorimetric detector. This system supports the growth of aerobic and anaerobic bacteria; bottles are available to support the growth of mycobacteria. This system has received Food and Drug Administration (FDA) clearance for monitoring bacterial contamination of platelets.
The BACTEC continuous-monitoring system (BD Diagnostics) is based on fluorescent technology. Bonded to the base of each vial is a CO 2 sensor that is impermeable to ions, medium components, and blood but freely permeable to CO 2 . If organisms are present, they release CO 2 into the medium; it then diffuses into the sensor matrix and generates hydrogen ions. The subsequent decrease in pH increases the fluorescence output of the sensor, changing the signal transmitted to the optical and electronic components of the instrument. The computer generates growth curves, and data are analyzed according to growth algorithms. Inoculated bottles are placed in individual cells of the instrument and rocked continuously. Adult and pediatric aerobic and anaerobic bottles are available, as well as a bottle for the recovery of yeasts and mycobacteria.
The VersaTREK system (Trek Diagnostic Systems) detects growth of organisms in broth by measuring gas consumption and gas production. Each inoculated vial is fitted with a disposable connector that contains a recessed needle. The needle penetrates the bottle stopper and connects the bottle headspace to the sensor probe. The sensor monitors changes within the headspace in the consumption and production of all gases (CO 2 , N 2 , and H 2 ) by growing organisms and creates data points internally in the computer. Media are available to identify aerobic and anaerobic bacteria, including mycobacteria. Some clinicians continue to request extended incubation protocols for Brucella spp. and other bacteria known as the HACEK group ( Haemophilus , Aggregatibacter [ formerly Actinobacillus] , Cardiobacterium , Eikenella , Kingella spp.). These organisms needed prolonged incubation times when manual methods predominated, but they can be detected during the routine incubation times of the automated instruments.
When mycobacteria or fungi are suspected, liquid medium developed by the manufacturer of automated and semiautomated broth culture systems can be used. These blood cultures are incubated for a prolonged period of 4 to 8 weeks.
Positive blood cultures containing commonly isolated aerobic organisms are usually detected within 12 to 36 hours of incubation. Until recently, the initial report was limited to a Gram stain; identification and susceptibility results could be expected no sooner than 24 to 48 hours after the Gram stain report. Both the FilmArray blood culture identification panel (BioFire Diagnostics) and the Verigene system (Nanosphere) use molecular methods to identify more than 90% of the organisms in the time when a Gram stain would be reported. They can also identify the presence or absence of mecA and other resistance genes, allowing empiric therapy to be tailored ( ). The Accelerate Pheno system (Accelerate Diagnostics) can identify the organisms causing 90% of bacteremias and provide minimal inhibitory concentrations of key antimicrobial agents within 8 hours of a positive blood culture ( ). As these systems evolve and continue to provide even more rapid, clinically actionable information, laboratories should work with clinical colleagues to make sure these more expensive modalities have impact on patient care. “Pushing” information to clinicians has been shown to decrease the time to escalation or de-escalation of empiric antimicrobials ( ).
The use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has shortened the time to identification. Although most laboratories allow for overnight incubation before testing mature colonies, an aliquot of the positive culture can be centrifuged and the pellet inoculated onto prewarmed plates. About 95% of isolates can be identified with the MALDI-TOF after 4 hours of incubation ( ). Although more labor intensive, the reagents of this method are much less expensive than the molecular methods. Cultures containing anaerobes are usually not detected for 48 to 72 hours, and identification is not available for 3 to 4 days. Fastidious organisms, such as those found in the HACEK group, may not be detected until 3 to 5 days.
In a small percentage of cases, no organisms will be seen on a Gram stain from a positive blood culture. It is very important to inoculate and monitor media in these cases. Brucella spp. can be difficult for an inexperienced eye to note on Gram stain; the colonies are visible on solid media a few days later. Conversely, some organisms are visible on Gram stain but not on routine media. These organisms grow on chocolate agar or sheep blood agar with a Staphylococcus aureus streak.
With regard to viruses, blood specimens are most commonly collected to monitor response of infection with human immunodeficiency virus (HIV), hepatitis C virus (HCV), hepatitis B virus (HBV), or cytomegalovirus (CMV) to antiviral therapy by using quantitative polymerase chain reaction (PCR) to measure viral load. Such assays are commercially available for each of these viruses, and in all cases, manufacturer’s guidelines for specimen collection and transport should be followed. For HIV and HCV, blood specimens also may be collected for genotyping (commercial assays are available), and PCR (qualitative or quantitative PCR) generally is used to confirm an initial positive HCV antibody result. As with viral load, manufacturer’s guidelines regarding specimen collection and transport should be followed.
In addition to assessing response to antiviral therapy, measuring viral load in a blood specimen is useful for monitoring disease and for diagnosis of disease in specific situations. In immunocompromised patients, especially transplant recipients but also patients with acquired immunodeficiency syndrome (AIDS), determining the level of CMV deoxyribonucleic acid (DNA) in blood is used to predict those at high risk for development of CMV disease and direct the initiation of preemptive therapy. Monitoring the level of BK virus and Epstein-Barr virus (EBV) DNA in serum or plasma by quantitative PCR is indicated in transplant recipients. If a commercially available NAAT assay is used, the package insert guidelines for specimen collection and transport should be followed. If, on the other hand, an assay developed and validated in house is used, guidelines published by that laboratory should be followed.
Blood specimens are useful for diagnosis of malaria, babesiosis, trypanosomiasis, and some filariasis ( ). Specimens should be collected in tubes with anticoagulant and transported promptly to the laboratory. If smears must be sent to a reference laboratory, they should follow the reference laboratory’s instructions for fixation soon after they are made. The techniques used in the laboratory for detecting the aforementioned parasites are the same and are discussed here in order of the simplest to the most complicated.
Standard Precautions need to be used when preparing smears and reading fresh (unfixed) smears. Additional fixation time in methanol could be recommended depending on the patient’s travel history. The simplest technique for detecting parasites in a sample of blood is the direct mount, prepared by placing 1 drop of blood on a glass slide, covering it with a cover glass, and examining it immediately. Direct mounts are excellent for diagnosis of trypanosomiasis or filariasis because the trypomastigotes and the microfilariae easily can be seen moving, often with low or medium power. Stained smears make the definitive diagnosis.
The thin smear, made as for hematologic work and stained in a similar manner, is the standard preparation for speciating Plasmodium spp., Babesia spp., Trypanosoma spp., and microfilaria. Thin smears for parasitologic work are fixed and then preferably stained manually with Giemsa stain, but automated hematologic staining is adequate. Smears are first scanned at low power to detect microfilariae, which are large objects (between 100 and 200 μm) and easily seen, usually at the lateral edges of the smear. After they are located, microfilariae should be studied under oil immersion for identification. After scanning with low power, the smear is examined with a high dry objective, searching for trypanosomes, and finally under oil immersion to find and identify Plasmodium, Babesia, and Trypanosoma spp.
Thick smears are useful for detecting all the parasites mentioned earlier and are part of the minimum laboratory workup for their diagnosis. One drop of blood is placed on a clean glass slide and, with the corner of another slide, is gently spread to cover 1 cm square. The preparation is allowed to dry and without fixation is stained with Giemsa stain, allowing for its dehemoglobinization.
CSF is collected to diagnose meningitis and, less frequently, viral encephalitis. Infectious meningitis, a medical emergency requiring early therapy to prevent death or serious neurologic sequelae, is divided into acute, subacute, and chronic clinical syndromes, based on duration of symptoms. Potential pathogens are listed in Table 66.1 . Enteroviruses are the agents most commonly responsible for meningitis, and they should be considered first in the differential diagnosis of meningitis in a child or adolescent during the late summer and early fall. The pyogenic bacteria responsible for meningitis vary with the age of the affected individual ( Table 66.2 ).
| Syndrome | Onset or Duration | Probable Pathogens |
|---|---|---|
| Acute | <24 hours | Pyogenic bacteria |
| Subacute | 1–7 days | Enteroviruses, pyogenic bacteria |
| Chronic | Persisting at least 4 weeks | Mycobacterium tuberculosis |
| Treponema pallidum | ||
| Brucella spp. | ||
| Leptospira interrogans | ||
| Borrelia burgdorferi | ||
| Cryptococcus neoformans | ||
| Coccidioides immitis | ||
| Histoplasma capsulatum |
| Age | Organisms |
|---|---|
| Neonates–3 months | Group B streptococcus |
| Escherichia coli | |
| Listeria monocytogenes ∗ | |
| Streptococcus pneumoniae | |
| 4 months–6 years † | Streptococcus pneumoniae |
| 6–45 years | Neisseria meningitidis |
| Older than 45 years | Streptococcus pneumoniae |
| Listeria monocytogenes | |
| Group B streptococcus |
∗ May cause meningitis in immunocompromised individuals in all age groups.
† Incidence of meningitis caused by Haemophilus influenzae type b in the United States has declined dramatically as a result of vaccination.
CSF is usually obtained by lumbar spinal puncture, but sometimes it is aspirated from the ventricles or collected from a shunt. As when collecting blood for culture, careful skin antisepsis is essential for collection of CSF, which typically is submitted to the laboratory in three or occasionally four tubes. The first tube filled should not be sent to Microbiology because it can contain skin flora. At least 0.5 to 1.0 mL should be sent to Microbiology for Gram stain and bacterial culture; ideally, the microbiology laboratory would receive 5 to 10 mL to accommodate requests for NAAT and cultures for mycobacteria and fungi ( ). The parameters of normal CSF and the usual changes that occur during meningitis caused by different organisms are listed in Table 66.3 .
| Condition | WBCs (cells/μL) a | Protein (mg/dL) | Glucose (mg/dL) |
|---|---|---|---|
| Normal | 5 (lymphocytes) | 14–45 | 45–100 (2/3 serum) |
| Meningitis | |||
| Acute or subacute bacterial | >500 (PMNs) | ↑ | ↓ |
| Chronic bacterial | 200–2000 | ↑ | ↓ |
| Tuberculous, fungal | (lymphocytes) | ↑ | ↓ |
| Enteroviral | 200–2000 (PMNs early; lymphocytes later) | ↑ | Normal |
CSF should be transported promptly to the laboratory and processed as rapidly as possible. If a brief delay in processing is unavoidable, the specimen should be held at room temperature unless viral culture is requested, in which case a portion (preferably 1 mL but no less than 0.5 mL) may be refrigerated for a short time. Specimen processing differs for bacteria, fungi, viruses, and parasites and is discussed separately for each group of organisms.
Processing CSF for routine bacterial culture includes concentration (if 1 mL or more of specimen is received), preparation of a smear by cytocentrifugation for staining with Gram stain, and culture. The supernatant is decanted into a sterile tube, leaving about 0.5 mL of sediment and fluid, which is thoroughly mixed on a vortex mixer or by forcefully aspirating up and down into a sterile pipette.
Diagnosis of chronic bacterial meningitis requires specific requests because the CSF is handled differently for each entity. To diagnose brucellosis, the CSF is processed as described earlier for routine bacterial culture, but the media are incubated for 2 to 3 weeks. For leptospirosis, Leptospira interrogans may be cultured from the CSF during the first few weeks of illness, but the Centers for Disease Control and Prevention (CDC) can test using a NAAT. The diagnosis of neurosyphilis can be challenging and requires neurologic signs and symptoms, reactive serologic results, and CSF findings including pleocytosis, elevated protein, and a positive Venereal Disease Research Laboratory (VDRL) test result. If the CSF-VDRL result is negative in the presence of clinical and serologic support for neurosyphilis, a CSF fluorescent treponemal antibody absorption test might be useful. This test is more sensitive and less specific that the CSF-VDRL ( https://www.cdc.gov/std/tg2015/syphilis.htm ). The specimen should be refrigerated until it is tested. Involvement of the central nervous system by Borrelia burgdorferi (Lyme disease) also is diagnosed serologically by detection of specific immunoglobulin (Ig) M and IgG antibodies in CSF and serum.
At least 5 mL of CSF is recommended for mycobacterial culture; cytocentrifugation is required to make a smear and optimize the yield. A NAAT may be useful, but a negative result does not exclude mycobacterial infection.
Processing CSF for detection of fungi is similar to that described for detecting bacteria. Organisms are concentrated by filtration or by centrifugation. A cytocentrifuge preparation or a smear of the sediment stained with Gram or other stain is examined, and appropriate media (e.g., brain-heart infusion or SABHI agar without antibiotics) are inoculated for culture.
In addition to culture, there is a commercially available molecular panel for meningitis including bacterial, viral, and fungal targets. This panel does not replace culture; the panel does not detect all CSF pathogens, and culture is still necessary for susceptibility results ( ; ).
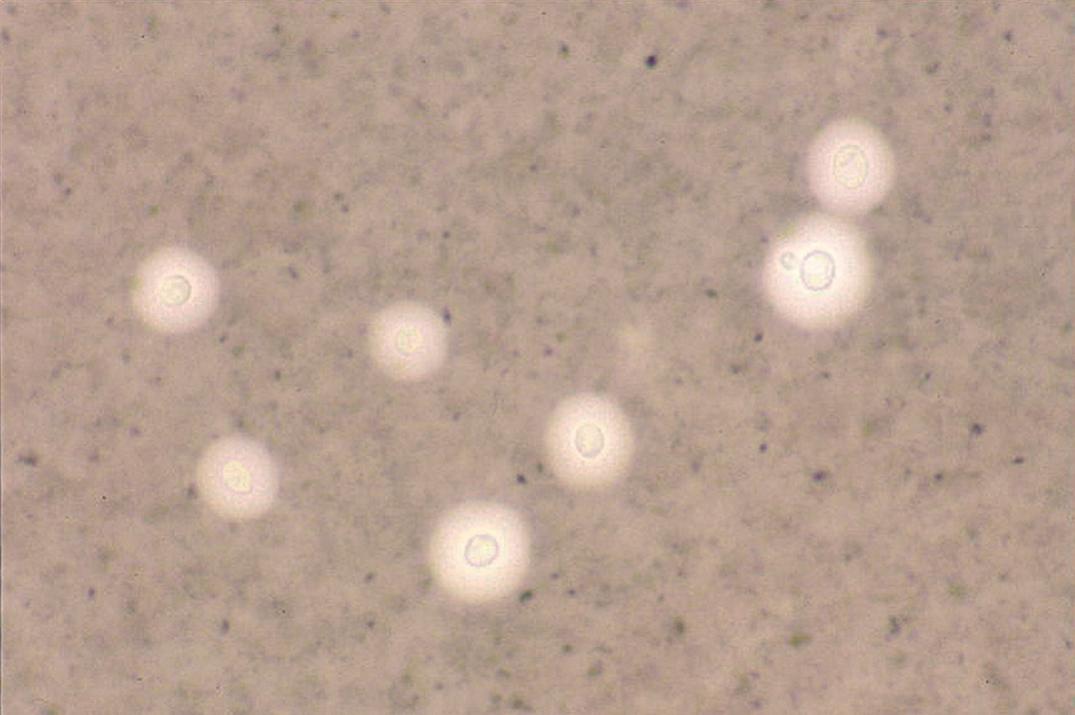
Two types of rapid tests are available for diagnosis of meningitis caused by C. neoformans: those specific for the capsular antigen (latex agglutination and enzyme-linked immunosorbent assay [ELISA]) and the nonspecific India ink preparation, which allow visualization of encapsulated yeast cells ( Fig. 66.1 ). The sensitivity of the India ink stain, performed by mixing 1 drop of CSF sediment with 1 drop of India ink (available at art supply stores), is low, except in HIV-infected persons. Therefore, the cryptococcal latex agglutination test or the ELISA, both of which are highly specific and have sensitivities of more than 90%, is recommended for diagnosis. Supernatant of a centrifuged specimen or unspun CSF can be used for these latter two tests.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here