Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
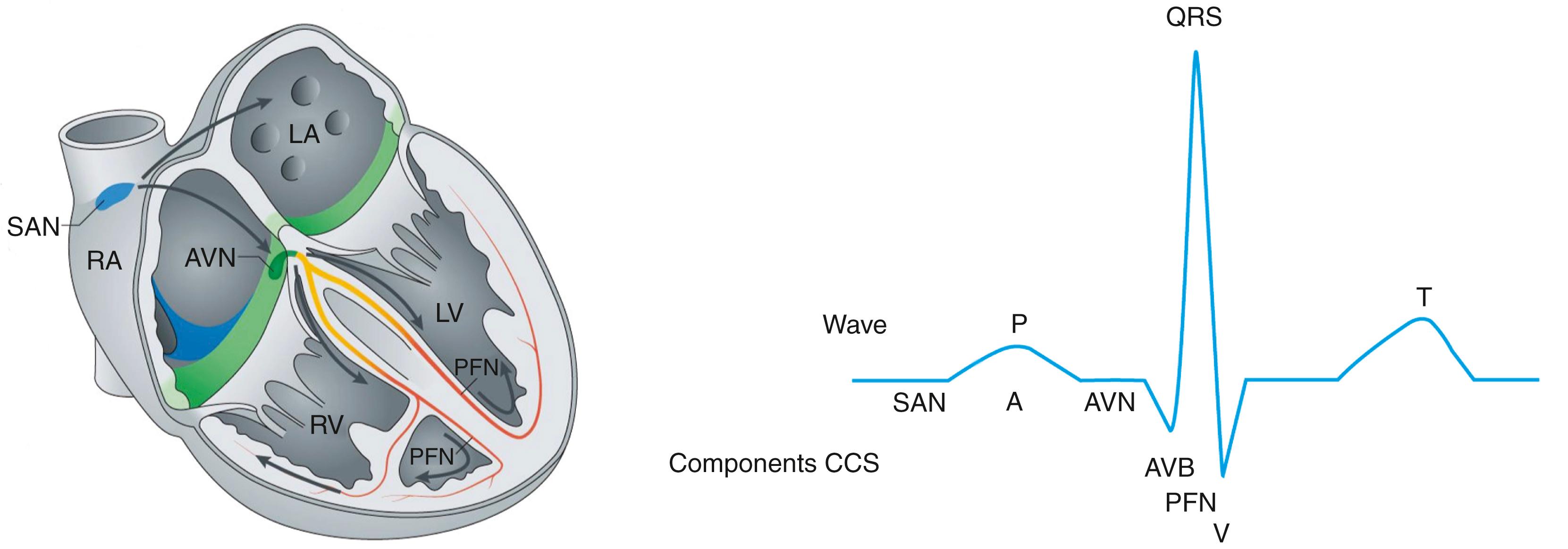
The electrical activity pattern of the mammalian heart, which depends on coordinated generation and propagation of action potentials, is essential for proper rhythmic contraction of the heart. Action potentials are generated and propagated by the cardiac conduction system (CCS) and working myocardium. The electrical impulse originates from the pacemaker cells in the sinoatrial node (SAN) followed by rapid propagation through the atria to the atrioventricular node (AVN). The AVN delays the propagation of the electrical impulse that allows atrial excitation and ventricular filling to complete before contraction of the ventricles. Next, the electrical impulse propagates through the fast conducting atrioventricular bundle (AVB), bundle branches (BBs), and Purkinje fibers to the ventricles, where the ventricular myocardium depolarizes, resulting in excitation of the ventricles ( Fig. 12.1 ). In addition to these differences in conduction velocities, the different types of cardiomyocytes also display differences in action potential shape, duration, and in diastolic membrane potential. Together, differences in tissue composition and architecture and in ion channel expression levels underlie the heterogeneous electrophysiologic properties of the mammalian heart. , Changes in expression or function of ion channels because of mutations that are linked to various inherited cardiac disorders (e.g., long QT syndrome, Brugada syndrome, atrial fibrillation, sinus node dysfunction) can give rise to potentially life-threatening arrhythmias, heart failure, and sudden cardiac death. Therefore knowledge of the genetic, genomic, and transcriptional mechanisms that regulate functional properties and expression of various ion channels is crucial in understanding the pathophysiology of heritable arrhythmias.
Genome-wide association studies (GWASs) have identified loci associated with electrocardiographic parameters such as PR interval and QRS duration. These GWASs demonstrate that the majority of disease- and trait-associated variants are found in noncoding regulatory elements of the genome in close proximity to ion channels (e.g., SCN5A-SCN10A, KCNQ1, KCNH2, HCN4 ) and cardiac transcription factors (e.g., TBX3-TBX5, NKX2-5, MEIS1, TBX20, HEY2 ). In addition, transgenic animal studies have uncovered a key role of several transcription factors in cardiac electrical activity. Together, these observations delineate a transcriptional regulatory network that regulates the spatiotemporal expression pattern of cardiac ion channels in mammals including humans. The T-box transcription factor Tbx5 is broadly expressed in the heart and is a transcriptional activator that promotes cardiac differentiation. , TBX5 regulates multiple genes involved in the development and maturation of the CCS. , Mutations in human TBX5 underlie the Holt-Oram syndrome, which is characterized by congenital heart defects and limb malformation, and conduction system dysfunction. Deletion of Tbx5 in the mouse ventricular conduction system results in a drastic decrease in Scn5a (Na V 1.5) and Gja5 (Cx40) expression; consequently, this leads to loss of fast conduction, arrhythmias, and sudden death. Tbx3 is selectively expressed in the CCS (SAN, AVN, AVB, and BBs) and controls the pacemaker gene program within its expression domain through a transcriptional repressive manner. Loss of Tbx3 causes embryonic lethality in mice. Nevertheless, reduction of Tbx3 within the heart has been associated with a wide spectrum of rhythm disturbances and lethal arrhythmias. Tbx3 and Tbx5 share a common DNA-binding domain in regulatory elementsm, , suggesting that these T-box transcription factors compete for binding and thereby tightly control spatiotemporal expression of target genes to regulate CCS function. Tbx3 and Tbx5 are dose-sensitive regulators. Alteration in the expression of either because of genetic variations in their regulatory elements can compromise the Tbx3/Tbx5 balance and the gene regulatory network for CCS development and function. Understanding the transcriptional regulation of cardiac rhythm and the physiologic role of regulatory elements is important to elucidate the exact mechanisms underlying the effects of disease- and trait-associated variants on cardiac conduction and arrhythmias. Tbx5 and Pitx2 were found to positively and negatively control downstream target genes essential for atrial rhythm including Scn5a and Gja5, but also calcium handling genes including Ryr2 and Atp2a2 (Serca2a) . Moreover, a subset of identified genes were previously associated with atrial fibrillation. This study reveals a transcriptional gene regulatory network maintaining atrial rhythm and provides a model to understand important aspects of the molecular pathophysiology of atrial fibrillation.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here