Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Genetic diseases are a leading cause of neonatal morbidity and mortality. Estimates of the incidence of genetic disease in neonates admitted to intensive care units have historically been based on targeted genetic testing of cohorts selected to have a high pretest probability, such as those with multiple congenital anomalies. However, the neonatal presentation of many thousands of genetic diseases overlaps with common, typically nongenetic conditions such as sepsis, hypoxic ischemic encephalopathy, and in utero infections. It is now possible to explore the potential for many nonspecific, life-threatening neonatal presentations (e.g., seizures, hypotonia, respiratory and cardiac failure) to have a monogenic or genomic etiology. This has become possible with the availability of clinical grade genome-wide sequencing technologies (whole exome sequencing [WES] or whole genome sequencing [WGS]). A recent study from regional neonatal intensive care units (NICUs) in the United States estimated that the minimum incidence of genetic disease is 14%, and highlighted the benefits of ultra-rapid diagnostic testing in a sub-set of neonates who were gravely ill and in whom individualized management may change the outcome. This study added to the increasing body of evidence to support the use of WES or WGS as a first-line test for critically ill neonates, in whom a genetic condition is suspected. , However, optimizing the clinical utility of such tests is dependent on healthcare providers understanding their strengths and limitations, as well as defining which patients will benefit, and at what point the test should be integrated into their care.
Healthcare policy makers worldwide are beginning to make decisions about the availability of, and eligibility for, neonatal genomic tests. It has never been more important for healthcare professionals who care for critically ill neonates to become familiar with the scientific rationale for the use of these evolving genomic technologies, as well as the ethical issues surrounding their implementation.
Genetic disease presenting in the neonatal intensive care unit represents the severe end of a spectrum of human genetic disease. Gains or losses of chromosome material, ranging in size from microdeletions to aneuploidies, affect the function of multiple genes required for health and development and have a tendency to present as severe prenatal or neonatal phenotypes. Because of this, the International Standard Cytogenomic Array (ISCA) Consortium recommends that a chromosomal microarray be used as a first-tier clinical diagnostic test in individuals with congenital anomalies or developmental disabilities. One study of chromosome microarrays in 638 neonates with birth defects found that 2.5% had aneuploidies and 12.7% had other copy number variants (CNVs). In recent years it has become possible, but is not yet routine, to detect CNVs by WES or WGS. A small but highly impactful number of diagnoses of CNVs are found in neonates in most large WES/WGS studies, , , even though, in many centers, sick neonates would have had a chromosome microarray prior to consideration for recruitment to these studies.
With regard to genomic sequence variants, there is a preponderance towards de novo autosomal dominant and autosomal recessive disorders in neonates. As many neonatal genetic disorders affect life span and reproductive fitness, a relatively low proportion of diagnoses are likely to be explained by inherited autosomal dominant disorders with similar phenotypes across multiple generations. In large studies of whole genome or exome sequencing in neonates, , , 46% to 84% of genetic diagnoses were autosomal dominant, 11% to 42% autosomal recessive, and 5% to 15% X-linked. In these studies, the majority (66% to 100%) of the autosomal dominant disorders were de novo rather than inherited. When a dominant genetic condition is proven to have occurred de novo in an affected neonate, there will be no family history consistent with similarly affected family members. However, there may be a history of older paternal age at conception, due to the mechanism discussed in the later section “Origin of Single Nucleotide Variants.”
A similar pattern of enrichment for de novo dominant disorders is seen in larger cohorts of infants with developmental disorders, perhaps with a lower rate of diagnosis of autosomal recessive disease than in neonates. In a trio WES study of 6040 families of children with developmental disorders (Deciphering Developmental Disorders), an estimated 42% of cases can be explained by de novo coding mutations, and just 3.6% by autosomal recessive disease (in patients with European ancestry). ,
Some autosomal dominant disorders exhibit remarkable differences in degrees of penetrance or expression, where the same pathogenic variant can leave the parents relatively healthy but the neonate very sick. In such instances, a detailed personal medical history and examination of the parents with additional enquiry regarding the family history can be critical.
For example, pathogenic variants in the SCN5A gene cause Brugada syndrome. This can predispose an individual to early-onset dangerous arrhythmias, whilst others with the same variant can remain free of arrhythmias until adulthood or for their entire lifetime. The family history in this instance may be as subtle as relative or relatives who drowned or had an unexplained road traffic accident. Another example is NOTCH1 pathogenic variants, which predispose to congenital heart disease (CHD). Penetrance is variable and can range from severe neonatal disease through to completely healthy adults. In this case the phenotype is predominantly congenital and, if survived, will usually result in a healthy adult life with reproductive fitness.
It is unclear why there can be such a wide spectrum of severity for many genetic disorders, even from the same pathogenic variant within the same family. The likelihood is that other genetic and environmental factors are acting to modify the phenotype.
Many pathogenic genetic variants in neonates arise de novo rather than being inherited from a parent. The mechanism by which these copying errors arise when a sperm or oocyte is being formed are linked to parental age.
Aneuploidy is present in greater than 35% of spontaneous abortions, 4% of stillbirths, and around 0.3% of live-born neonates, reflecting the severe phenotypes and embryonic lethality of many of the aneuploidies.
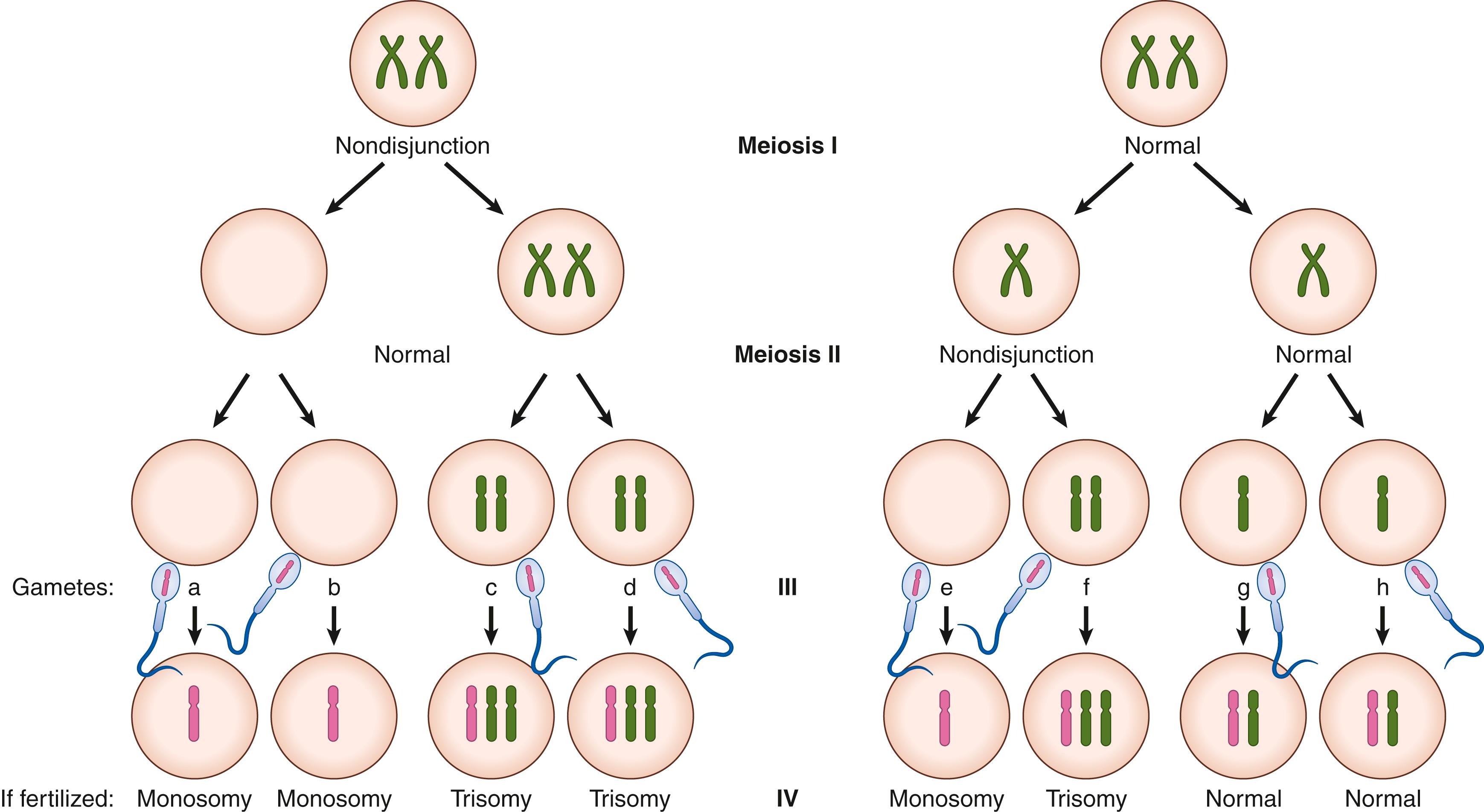
Aneuploidy originates during cell division when the chromosomes do not separate equally (nondisjunction). If this occurs in the germ cell lineage during meiosis, it can result in oocytes or sperm with an aberrant number of chromosomes and so result in an aneuploid embryo. The origin of aneuploidy is predominantly from oocytes. Around 10% to 35% of human oocytes and 1% to 4% of sperm are aneuploid. The incidence of aneuploidy increases with maternal age. The precise mechanism by which aging oocytes are prone to aneuploidy is unclear and likely multifactorial. Indeed, the relationship between maternal age and aneuploidy is different for different chromosomes, suggesting multiple pathways at play ( Fig. 173.1 ).
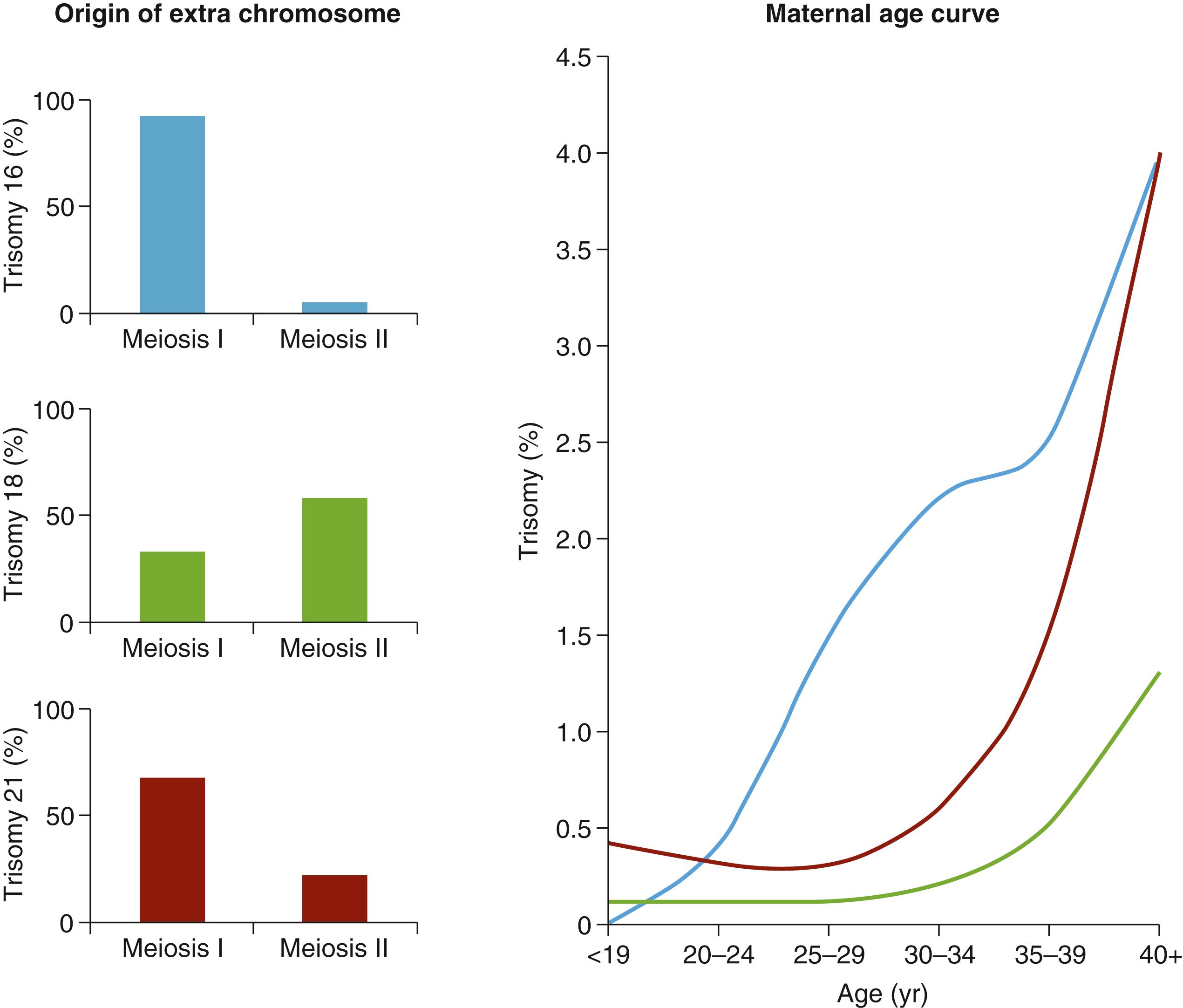
Nondisjunction can occur at the first or the second cell division of meiosis (meiosis I or meiosis II— Fig. 173.2 ). Oocytes have a fundamentally different mechanism of meiosis from sperm, which have symmetric cell divisions and proceed swiftly through meiosis. Oocytes are arrested in prophase I from embryonic development until puberty. At this point, luteinizing hormone stimulates the resumption of meiosis. At anaphase I, homologous chromosomes separate; one remains in the oocyte and the other forms the first polar body. The ovulated oocyte is arrested in metaphase II. Fertilization triggers the second meiotic division with separation of the sister chromatids: one in the oocyte and the other into a second polar body. As this process of a single meiotic division lasts decades in oocytes, this provides opportunities for segregation errors to occur.
The relative contribution of meiosis I errors and meiosis II errors in oocytes varies according to the chromosome in question, but in general meiosis I errors are more common in natural pregnancies —see Fig. 173.1 . Failure of meiotic recombination or suboptimal location of crossover points predisposes to aneuploidy, and aging oocytes suspended in meiosis suffer a loss of sister chromatic cohesion.
Embryos mosaic for aneuploidy can also form (with a mixture of aneuploidy and euploid cells). There is evidence that aneuploidy cells can suffer apoptosis and proliferative defects, meaning that the proportion of aneuploid cells becomes progressively depleted as the embryo develops. This happens—to a lesser extent—in placental tissue compared to fetal tissue, meaning that prenatal tests that sample placental DNA (including chorionic villus sampling and free fetal DNA sequencing) may overestimate the degree of mosaic aneuploidy in the fetus.
In contrast to aneuploidies, germline sequence variants predominantly originate from the sperm rather than oocytes, with a ratio of around 3.5:1. The rate of de novo genetic variants in sperm increases as a male ages. This is because, in contrast with oocytes, which are produced early in life and have a fixed number of replication cycles, sperm continue to undergo cell divisions throughout a male’s reproductive lifetime. Each cell division requires copying of the DNA and a chance for replication errors. Sperm have undergone approximately 160 replications in a 20-year-old man, and this increases to 610 in a 40-year-old man. Each sperm carries two to three additional de novo sequence variants per additional year of paternal age ( Fig. 173.3 ). ,
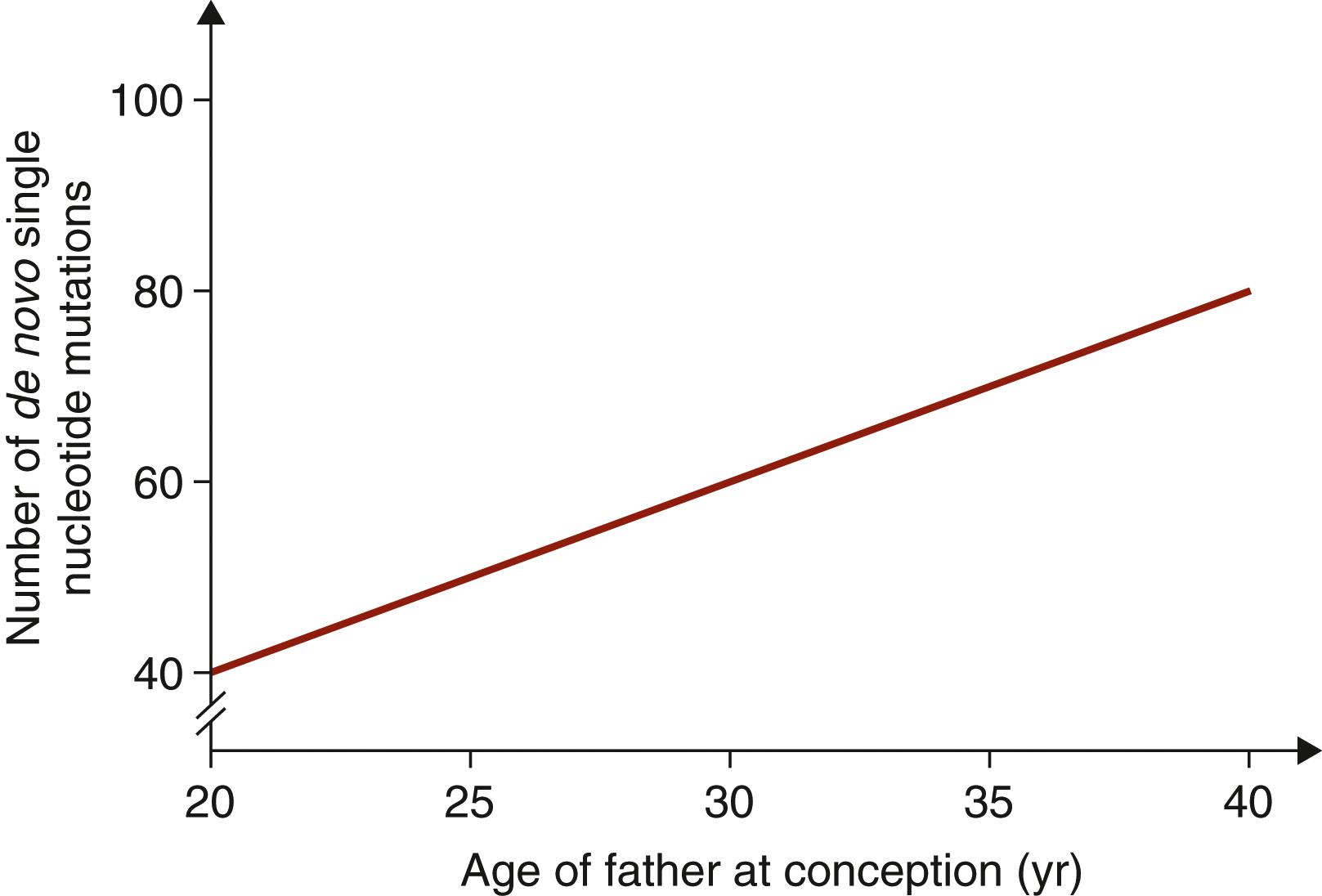
Some genetic variants give sperm cells a survival or proliferation advantage. Sperm with these variants will drift upwards in frequency as they form a rapidly dividing clone within the testis. An example is the FGFR2 p.Ser252Trp mutation, which is seen in 62% to 71% of cases of Apert syndrome. The frequency with which sperm are seen with this point mutation is far higher than would be expected due to chance as FGFR2 is a growth factor receptor and activating mutations will cause greater growth and proliferation of the sperm.
The origin of de novo large CNVs is currently less clear. Some studies have suggested a bias towards paternal origin of deletions, and a maternal origin of duplications. ,
CNVs are deletions or duplications of a large number (usually at least 1000) of DNA base pairs. CNVs have traditionally been detected using a chromosome microarray: a first-line genetic test in the context of multiple congenital anomalies. The ability of WGS/WES technologies to detect CNVs is improving, and more CNVs are likely to be diagnosed this way in the future, using WGS/WES as a combined test for both CNVs and sequence variants.
As CNVs usually disrupt the expression of a number of genes, they frequently have multiple phenotypes of early or congenital onset. The severity of the phenotype will depend on a number of factors, not only the size of the CNV but also its location. Some genomic loci are more gene-rich or contain genes more critical to health and development than others. Deletions tend to have a more severe phenotypic effect than duplications as many genes are “haploinsufficient”—a single copy of the gene is insufficient to produce enough protein product to allow the cell to function normally.
The most extreme example of CNVs are the aneuploidies—where there is a gain or loss of whole chromosomes. Most of these are embryonic lethal with a few notable exceptions: trisomies 13, 18, or 21, sex chromosome aneuploidies, and mosaic aneuploidies.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here