Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Human immunodeficiency viruses (HIV-1, HIV-2) cause progressive immune deficiency and death from opportunistic infections or neoplastic diseases. Increased understanding of HIV pathobiology has led to significant advances in diagnosis ( Chapter 355 ), prevention ( Chapter 356 ), and treatment ( Chapter 357 ) that have reduced morbidity and mortality, and tipped the balance toward control of the epidemic worldwide.
HIV is a member of the lentivirus genus of the Orthoretrovirinae subfamily of the Retroviridae family of Revtraviricetes. All retroviruses contain reverse transcriptase, which catalyzes the synthesis of DNA from an RNA template. Retroviruses are widely distributed among vertebrates; lentiviruses cause chronic, recurrent, or progressive diseases, including immunodeficiencies, in various mammalian species. Several cross-species transmissions of lentiviruses from primates to humans were the source of human immunodeficiency viruses ( Chapter 353 ).
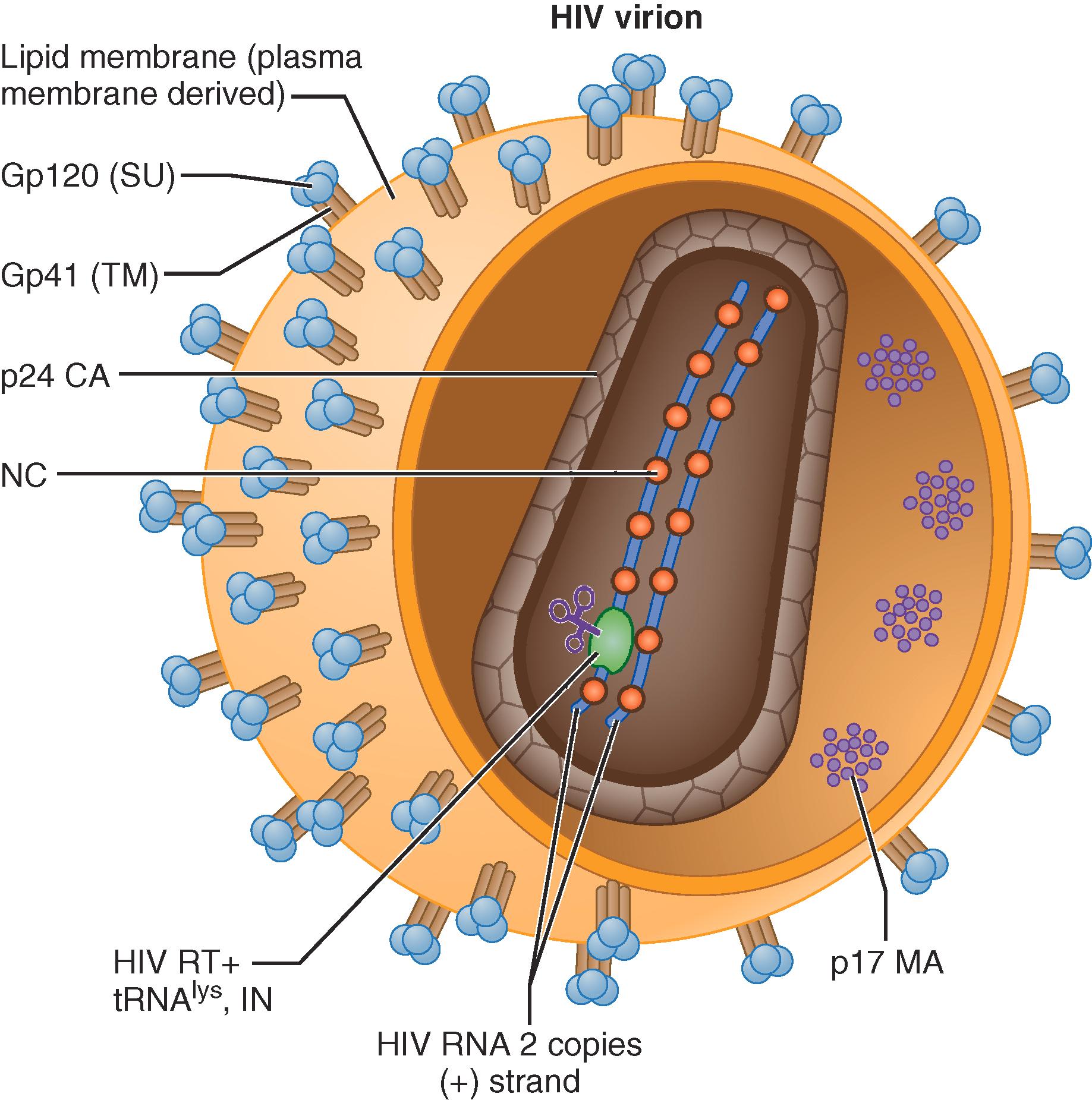
HIV virions are enveloped particles that are approximately 100 nm in diameter ( Fig. 354-1 ) and contain: two single-stranded (+) sense RNA copies, which code for HIV genes ( E-Fig. 354-1 ); the tRNA primer, within a virion protein core composed of structural proteins; the viral enzymes; reverse transcriptase and integrase essential for replication; and viral factors (Vpr and Vif) that are necessary to evade intracellular restriction. The viral envelope, which is derived from the host plasma membrane at budding, contains viral proteins Gp120 (SU) and Gp41 (TM), as well as a structural matrix protein, MA.
Viral replication ( E-Fig. 354-2 ) is initiated by direct contact of virions or infected cells with susceptible host cells ( E-Fig. 354-2A ) mediated by surface Env glycoprotein trimers of HIV sp120 SU noncovalently bound to HIV gp41 TM. Attachment is mediated by SU, which engages two distinct cell surface proteins, a receptor, and a coreceptor. HIV initially binds the CD4 receptor, thereby resulting in conformational change in Env and facilitating binding to a coreceptor, typically either the human chemokine receptor CCR5 or CXCR4 (although CCR5/CXCR4 dual-tropic viruses circulate as well). Engaging both receptor and coreceptor results in conformational change in TM to drive membrane fusion. Inhibitors of SU-CD4, SU-CCR5 interactions, and TM-mediated fusion are effective treatments for HIV.
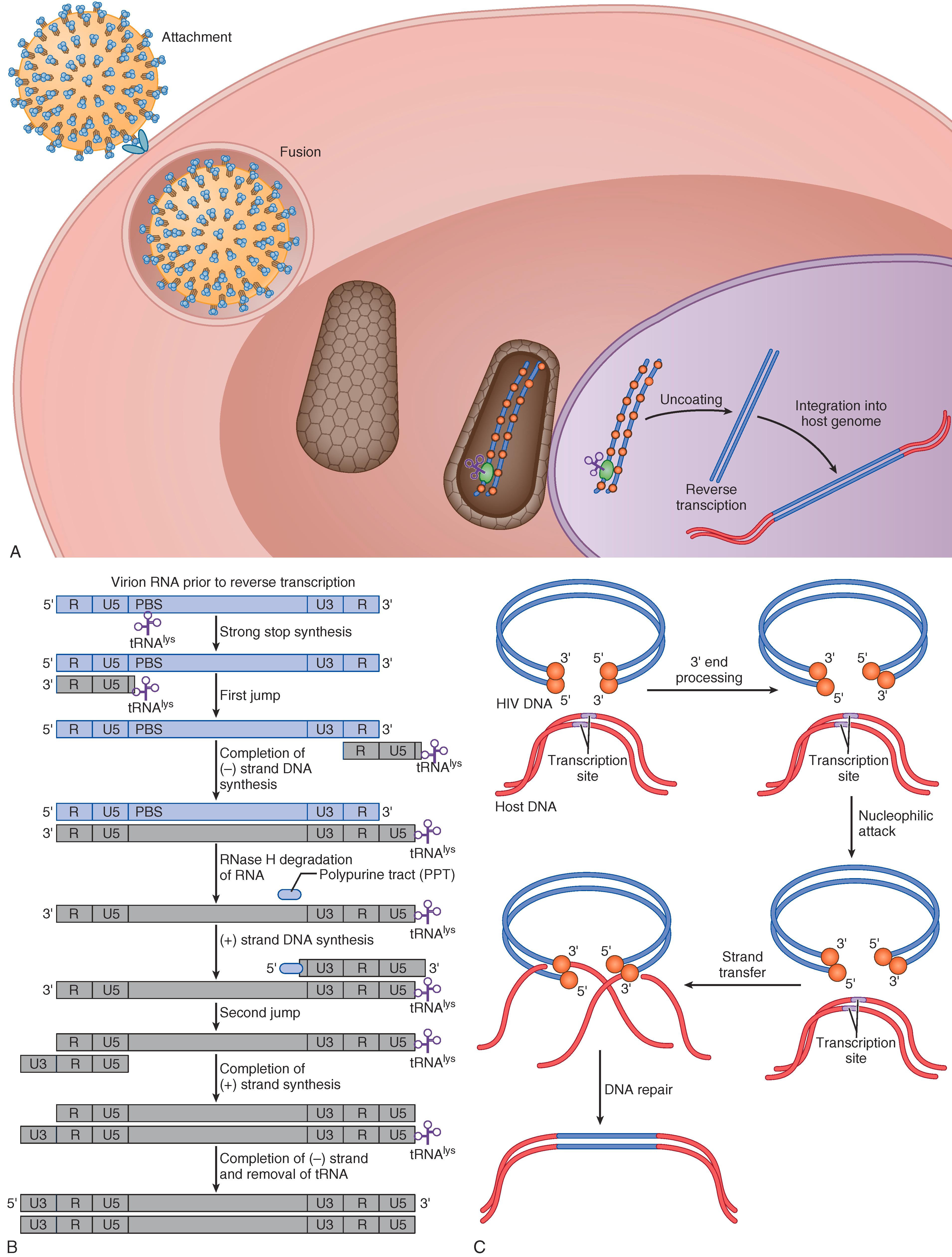
A key event in HIV replication is the conversion of virion RNA into double-stranded DNA by reverse transcriptase. Effective reverse transcription and uncoating occurs in the nucleus, followed directly by integration into the host’s genome ( E-Fig 354-2B and 2C ); potent inhibitors of reverse transcriptase, integrase, and uncoating are now FDA-approved ( Chapter 357 ).
Reverse transcriptase is a highly error-prone enzyme, with only a rudimentary editing function; approximately one mutation per virion occurs per replication cycle. The HIV generation time is estimated to be relatively short (1 to 2 days), and the replicating population size is substantial. This combination of rapid and error-prone replication results in a genetically diverse population that can respond rapidly to immune- or drug-selective pressure. Thus rapid error-prone replication represents a critical pathogenic determinant for HIV. Estimates indicate that variants with single resistance mutations to individual antiretroviral agents preexist in HIV-infected patients prior to initiating antiretroviral therapy. As a consequence, antiretroviral therapy requires combinations of drugs with distinct resistance profiles to prevent the emergence of resistance. Combinations that inhibit one to three steps in replication can be effective ( Chapter 357 ).
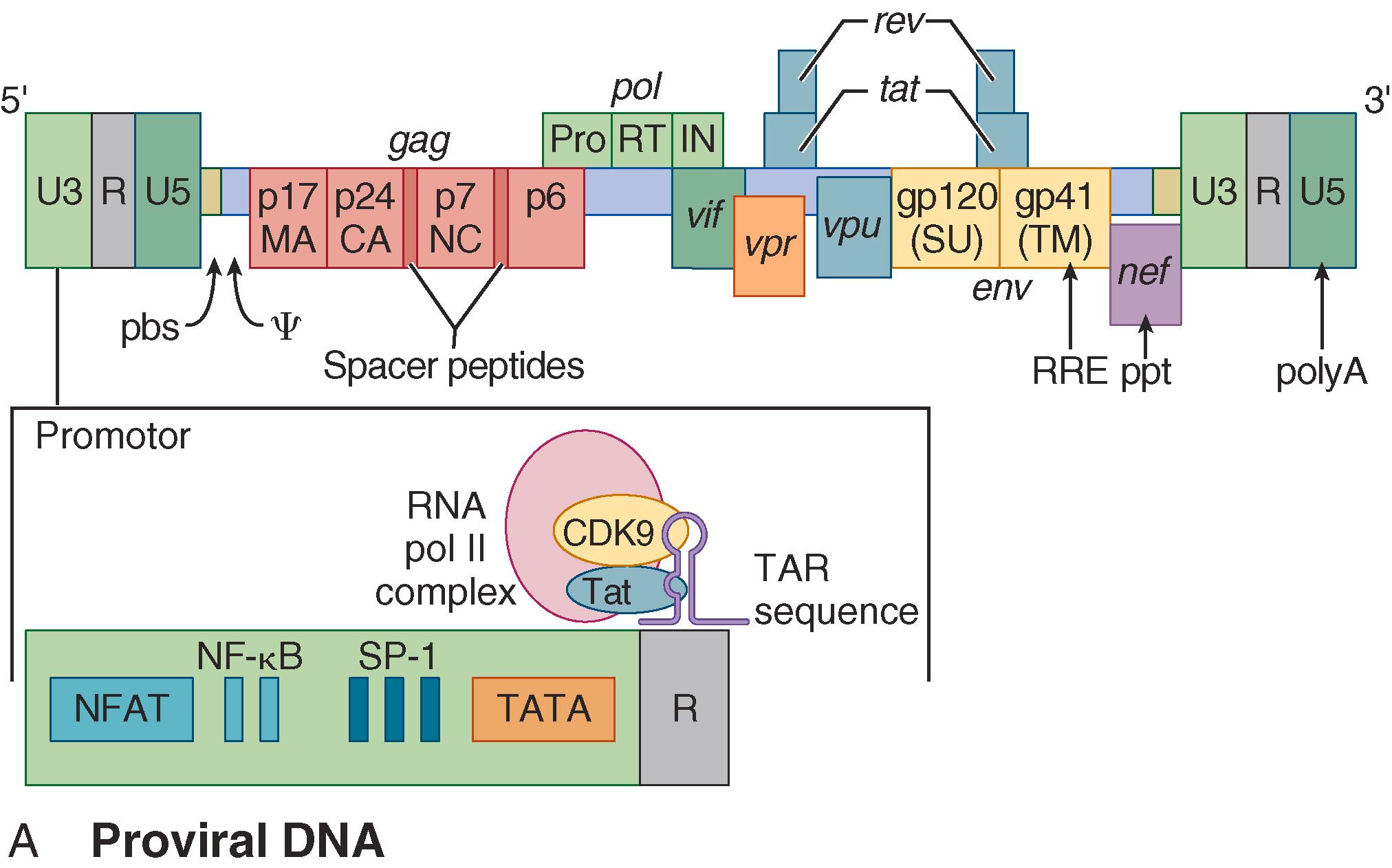
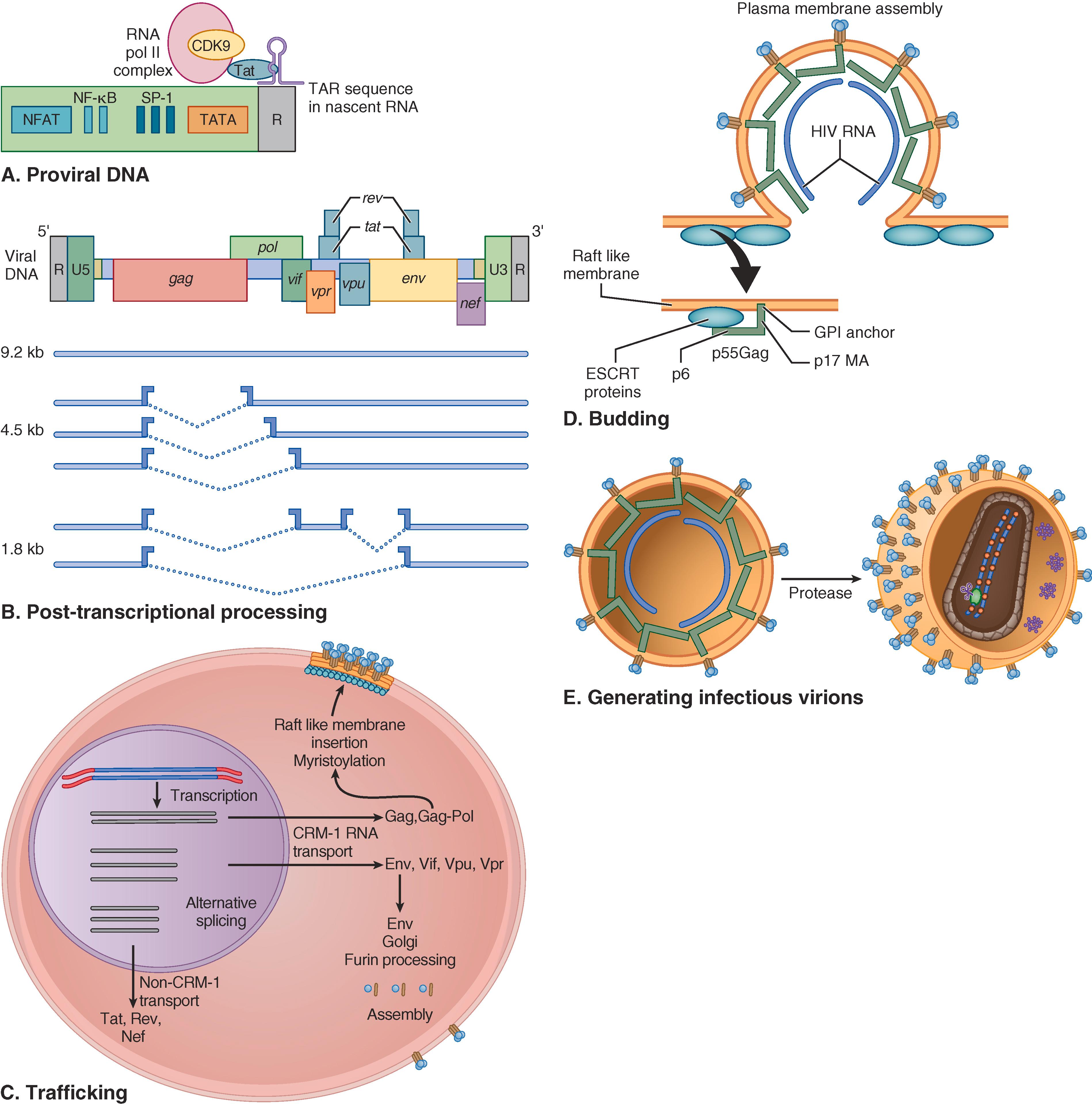
After the proviral state is established, HIV produces viral RNA and proteins using viral factors in concert with cellular mechanisms of transcription and translation. HIV replicates not by dismantling cell functions but rather by using specific interactions among host factors and specific viral gene products that drive high levels of HIV RNA transcription (Tat), facilitate balanced RNA transport (Rev), and help traffic of HIV proteins to membranes for virion budding (Gag) ( E-Fig. 354-3 ). Following budding, HIV Gag precursor proteins require proteolytic processing by the viral enzyme protease for mature core formation, which is required for viral infectivity. Potent protease inhibitors, which block this step, are important components of combination antiretroviral therapy ( Chapter 357 ). Maturation of HIV virions represents a new target for inhibition, and several agents are under active investigation.
HIV infection triggers a number of intracellular innate responses, which, if left unchecked, would arrest HIV replication. Several interferon-induced genes inhibit HIV replication, including the ABOBEC family of nucleic acid editing enzymes; the cell surface protein BST-2 (tetherin), which blocks release of virions from the cell surface; and SERINC 3/5, which incorporates into virion membranes and reduces infectivity of the produced virions. In each case, a viral factor (Vif for APOBEC, Vpu for BST-2, and Nef for SERINC 3/5) blocks these innate mechanisms by binding and redirecting corresponding host proteins to ubiquitin-induced degradation. Several HIV proteins (Vpiu, Nef) also down-modulate immune cell surface proteins CD4 and MHC molecules, thereby hindering adaptive immune responses.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here