Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Although ion channels and transporters are important modulators to mediate the ion flux across the plasma membrane in various cell types, including cardiomyocytes, they are also expressed on intracellular membranes to regulate various organellar and cellular functions. These intracellular membrane structures include the endoplasmic reticulum (ER)/sarcoplasmic reticulum (SR), Golgi apparatus, endosomes, lysosomes, mitochondria, nucleus, chloroplasts, and vacuoles. Among these organelles, the ion channels and transporters located at ER/SR have been studied extensively (as described in some of the other chapters). Indeed, two ER/SR-localized Ca 2+ release channels, inositol trisphosphate (IP 3 )-receptors and ryanodine receptors (RyRs), and an uptake transporter, SR Ca 2+ -ATPase (SERCA), have been a leading topic for organelle ion channel/transport research in the cardiovascular field. Studies have revealed that the mitochondrial ion channels and transporters also directly or indirectly affect cardiac electrophysiology in both physiologic and pathologic conditions via regulating intracellular Ca 2+ signaling, cellular oxidation, and bioenergetics. , This chapter covers the functional implications of mitochondrial ion channels and transporters in general and their impact on cardiac electrophysiology and pathology in particular.
Mitochondria are the so-called “power plants” of all kinds of tissues/cells. In the heart/cardiomyocytes, mitochondria use glucose and fatty acids to produce adenosine triphosphate (ATP), which drives muscle contraction and relaxation for pumping blood during each heartbeat. In the first half of the 20th century, mitochondria were studied mostly as cellular power plants. Soon it was also recognized that Ca 2+ stimulates oxidative phosphorylation (OXPHOS) and electron transport chain (ETC) activity, which results in the stimulation of ATP synthesis ( Fig. 7.1A–B ). Early studies in the 1960s and 1970s revealed that isolated mitochondria take up a large quantity of Ca 2+ . Surprisingly, superphysiologically high Ca 2+ concentrations ([Ca 2+ ]) (10–100 μM) were required to activate Ca 2+ uptake into isolated mitochondria (see reviews , ). In the intact cells, however, Ca 2+ was taken up by mitochondria during global increase of cytosolic [Ca 2+ ] ([Ca 2+ ] c ) (≤ 10 μM). This discrepancy between isolated mitochondria and intact cells was partially resolved by the finding of high [Ca 2+ ] c at the microdomains between mitochondria and ER/SR during Ca 2+ release via IP3 receptors (IP3Rs) and/or RyR because of the juxtaposition of these two organelles. These seminal discoveries have finally positioned mitochondria as one of the key players in the dynamic regulation of physiologic Ca 2+ signaling and promoted research in the fields of mitochondrial Ca 2+ channels/transporters. Soon it was also discovered that a Ca 2+ efflux mechanism exists to move the accumulated matrix Ca 2+ into the cytosol.
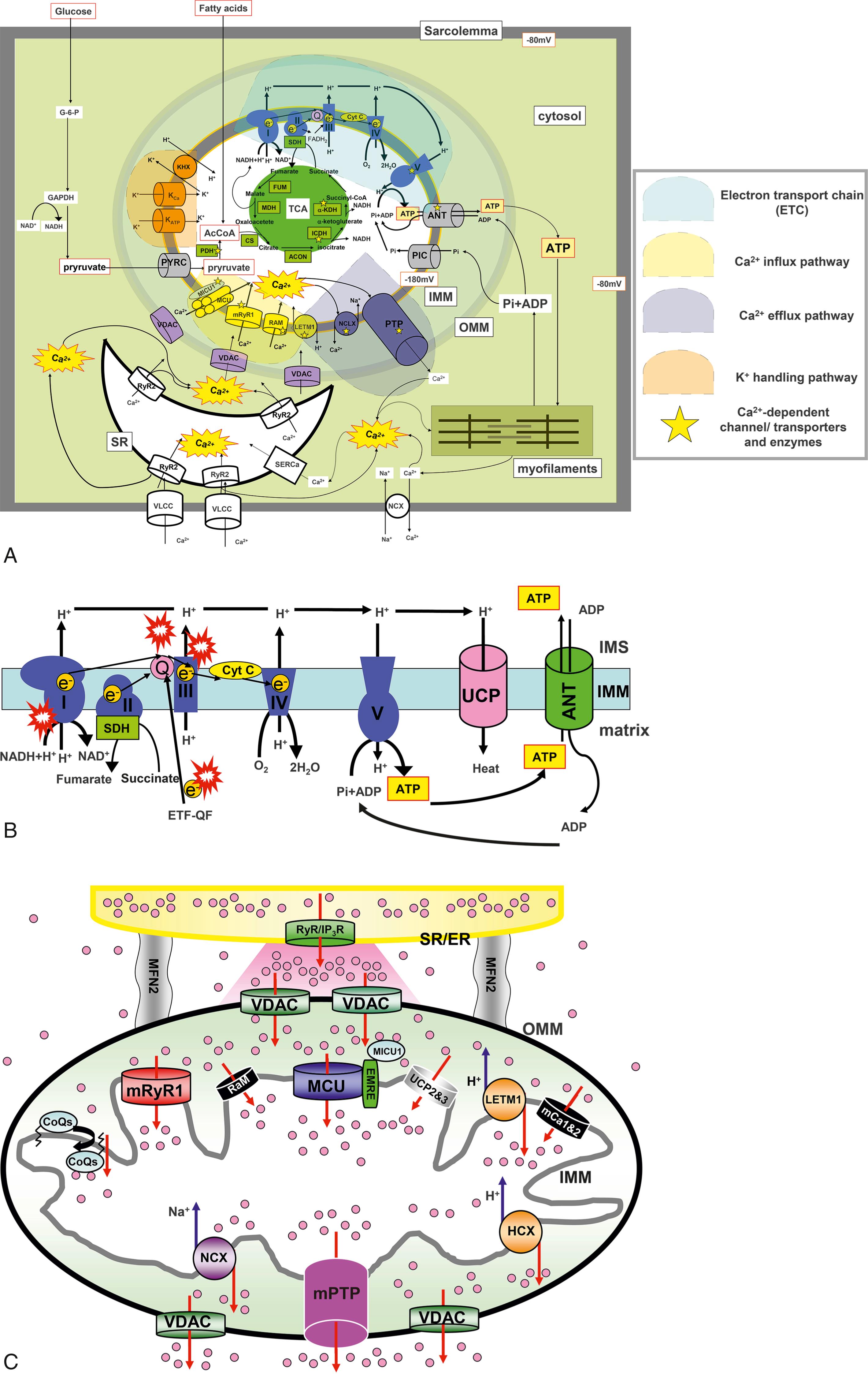
Although the functional characteristics of mitochondrial Ca 2+ influx/efflux mechanisms were discovered over 50 years ago, the molecular identities responsible for these mechanisms have remained a mystery until very recently. The mitochondrial Ca 2+ influx was dogmatically considered to result from a single transport mechanism mediated by a mitochondrial Ca 2+ uniporter (MCU) principally because of nearly complete inhibition by ruthenium red (RuR) and lanthanides, as was originally reported over 30 years ago. , Finally, groundbreaking studies have made progress, unveiling that MCU is encoded by CCDC109A . , These findings open up an exciting opportunity for investigating mitochondrial Ca 2+ handling by using genetic approaches. For instance, the molecular identification of MCU set a foundation for the discovery of several auxiliary subunits that associate with this pore-forming protein (e.g., EMRE and MICU1–3). , From these discoveries, it was conceptualized that uniporter activity is not solely regulated by one single protein (MCU) but possibly by macromolecular protein complexes (called mitochondrial Ca 2+ uniporter protein complexes or mtCUC ). , Furthermore, several groups showed that kinases in the cytosol can translocate into the mitochondria, interact with mtCUC, and directly modulate mtCUC function through posttranslational modifications (PTMs) of MCU in cardiomyocytes. These reports also support the idea that the MCU and its regulatory proteins form macromolecular local signaling complexes at the inner mitochondrial membrane (IMM), which may significantly affect mitochondrial Ca 2+ handling and numerous mitochondrial and cellular functions. Moreover, subsequent studies have identified additional Ca 2+ uptake pathways, which exhibit different Ca 2+ affinity, uptake kinetics, and pharmacologic characteristics from the original MCU theory (see reviews , ; see also Fig. 7.1C ). These distinct properties enable mitochondria to carry out multiple Ca 2+ -mediated functions with optimal spatial and temporal effectiveness. In cardiomyocytes, ryanodine receptor type 1 (RyR1) was the first mitochondrial Ca 2+ influx mechanism that was found to have a molecular identity, as reported by our group (see “Mitochondrial Ryanodine Receptor”). Through RNA interference studies, several groups have proposed other novel candidate proteins, in addition to our RyR1, that are involved in mitochondrial Ca 2+ uptake mechanisms, such as Letm1.
In addition to Ca 2+ , the movements of various electrolytes and metabolites across the IMM and the outer mitochondrial membrane (OMM) are important for the regulation of major mitochondrial functions including ATP synthesis, Ca 2+ homeostasis, and reactive oxygen species (ROS) and nitrogen species (RNS) generation. Unlike other organelles, mitochondria possess unique double-membrane structures with distinctive phospholipids and protein compositions, which allow mitochondria membranes to maintain their membrane potential (ΔΨm) and unique architecture, including cristae. Interestingly, the IMM and OMM have different sets of ion channels/transporters, as summarized in Fig. 7.1A . These include (1) proton (H + ) movement related to ETC activity and uncoupling proteins (UCPs) for the maintenance of ΔΨm and pH gradient (ΔpH) across the IMM; (2) K + -selective and anion-selective pathways at the IMM and across the two membranes, such as the mitochondrial permeability transition pore (mPTP), which are important for the maintenance of mitochondrial volume; (3) the movement of metabolites including ATP, adenosine diphosphate (ADP), and phosphate (Pi) at the IMM and OMM; and (4) large pores mainly at the OMM or mPTP that release proapoptotic proteins, which potentially lead to cell death. This chapter summarizes the progress in mitochondrial ion channel/transporter research and provides an overview of cardiac mitochondrial ion channel/transporter biophysics and cardiac physiology and pathophysiology.
The most prominent contribution of mitochondria to cellular metabolism is based on their capacity to generate ATP through the tricarboxylic acid (TCA) cycle and OXPHOS through the ETC. This is achieved by a concerted series of redox reactions catalyzed by four multisubunit enzymes embedded in the IMM (Complex I–IV) and two soluble factors, cytochrome c (cyt c) and coenzyme Q10 (Co Q10), which function as electron shuttles within the mitochondrial intermembrane space (IMS; see Fig. 7.1B ). In eukaryotic cells, more than 90% of the total intracellular ATP is generated by mitochondria. The main driving force of OXPHOS, known as chemiosmosis , is generated by the proton (H + ) movement across the IMM that creates a membrane potential (ΔΨm, negative in the matrix) and a pH gradient (ΔpH, alkaline in the matrix). Chemiosmosis is the movement of ions across a selectively permeable membrane, down their electrochemical gradient (proton motive force: Δp), which is determined by both ΔΨm, and ΔpH components across the IMM (Δp =ΔΨm + ΔpH). The chemiosmosis hypothesis was first proposed by Peter D. Mitchell in 1961. The basic assumption of this chemiosmosis theory is derived from the important observation that the IMM is generally impermeable to ions, but it keeps the permeability of H + . The composition of the OMM is similar to those of sarcolemma and ER/SR in eukaryotic cells, whereas the IMM does not possess cholesterol but has cardiolipin, a dimeric phospholipid that is typically found in bacterial membranes. Cardiolipin has a unique ability to interact with proteins, including several mitochondrial respiratory chain complexes (I, III, IV, and V), and support their activities but also contributes to maintaining the structure of cristae, which enhances the efficiency of ETC activity, possibly by facilitating the formation of super complexes of respiratory chain at the IMM. The unique structure of cardiolipin serves as a H + trap at the IMS near the IMM, maintains the pH change near the IMM, and efficiently pools H + or releases H + to the mitochondrial ATP-synthase (complex V) at the IMM (see Fig. 7.1B ). , Interestingly, the IMM and OMM not only have the different phospholipid compositions but also show different protein-to-lipid ratios (for OMM, about 1 and for IMM about 4). This may allow the proteins at the IMM to possess enzymatic and/or transport functions compared with those at the OMM. The IMM is much less permeable to ions and small molecules than the OMM, which provides the cellular compartmentalization between the mitochondrial matrix and cytosol. As shown in Fig. 7.1B , complexes I, III, and IV are engaging with the translocation of H + from matrix to IMS, which establishes ΔΨm, and ΔpH (i.e., Δp). Therefore, ΔΨm is usually highly negative (around −180 mV) compared with the resting potential at plasma membranes. This large driving force for H + influx (Δp) is finally used by complex V to produce ATP (see Fig. 7.1B ). There are other important roles for Δp, in addition to ATP synthase at the IMM: (1) ΔpH drives pyruvate transport through pyruvate carrier (PYRC) into the matrix; (2) ΔpH drives Pi transport through Pi carrier (PIC) into the matrix; and (3) ΔΨm drives the ATP/ADP exchange through the adenine nucleotide translocator (ANT; see Fig. 7.1A–B ).
As previously mentioned, Ca 2+ uptake into the mitochondrial matrix stimulates ATP synthesis (see Fig. 7.1A ). At the resting state, the electrochemical driving force for Ca 2+ uptake is also provided by ΔΨm across the IMM. For MCU, Ca 2+ is taken into the mitochondrial matrix down its electrochemical gradient without transport of another ion. Basically, for each Ca 2+ transported through MCU, there is a net transfer of two positive charges into the matrix that results in an energetically unfavorable drop of ΔΨm . The Ca 2+ -stimulated respiration, however, will not only compensate the loss of ΔΨm by the efflux of H + via ETC but also will produce a net gain of ATP. Indeed, a study from Boyman’s group showed that Ca 2+ influx via MCU in cardiac mitochondria may not directly stimulate mitochondrial bioenergetics; instead, changes in matrix [Ca 2+ ] ([Ca 2+ ] m ) guides ATP production by its influence on the ΔΨm . Finally, multiple Ca 2+ efflux mechanisms also work in concert to expedite a transient and oscillatory nature rather than a tonic and steady state change of [Ca 2+ ] m .
Superoxide and nitric oxide are key elements of ROS and RNS, respectively, produced in cells under normal physiologic conditions, and both species react with other molecules and each other to form a diverse array of additional ROS and RNS. High levels of ROS and RNS promote cell damage and death, but the production of low to moderate levels of ROS/RNS is critical for the proper regulation of many essential cellular processes, including gene expression, signal transduction, and cardiac excitation-contraction (E-C) coupling. ROS are generated by several different cellular sources: (1) membrane-associated NADPH oxidase, (2) cytosolic xanthine and xanthine oxidase, and (3) the mitochondrial ETCs at the IMMs.
Superoxide is the primary oxygen free radical produced in mitochondria via the slippage of an electron from the ETC to molecular oxygen during OXPHOS (see Fig. 7.1B ). This “constitutive” superoxide generation is central to the proper cellular redox regulation. Studies from our collaborating groups and other investigators detected a “stochastic” and “transient” superoxide and alkalization burst from either single or restricted clusters of interconnected mitochondria across a wide variety of cell types, termed mitochondrial flash. , A transient depolarization of ΔΨm is associated with each mitochondrial flash. The proposed mechanism for the mitochondrial superoxide flash (mSOF), a component of the transient superoxide burst in cardiac muscle cells, is as follows. A small increase in constitutive ROS production transiently opens a large channel called the mPTP to cause ΔΨm depolarization, which subsequently stimulates the ETC to produce a burst in superoxide production. This idea is similar to the previous observation that the mPTP opens and closes transiently (“flickers”) at its low conductance state and can release Ca 2+ from the matrix. , The frequency of mitochondrial flashes varies widely across different cell types and experimental conditions (disease models), suggesting that these flashes act as biomarkers of cellular metabolic activity and oxidative condition in physiologic and pathophysiologic conditions. , Future studies will clarify the contribution of altered mSOF activity to ROS overproduction and metabolic dysfunction in a wide range of mitochondrial diseases and oxidative stress related disorders. Finally, it should be noted that mitochondrial flashes are composed of a superoxide signal with an alkalization signal within the mitochondrial matrix. , Based on chemiosmosis theory, one can envision that whenever there is a significant drop in ΔΨm, there should be an alkalization in ΔpH. The functional role of this concurrent digitized superoxide and pH signal awaits further investigation.
Programmed cell death is genetically designed for the self-killing of individual cells and is one of the critical mechanisms for maintaining homeostasis of multicellular organ/tissues, including the heart. Apoptosis is a well-established mechanism of programmed cell death activated by a variety of cellular stresses and signals. In mammalian cells, the activation of caspases (a family of cysteine proteases) is the central mechanism for apoptosis. Under the resting condition, caspases are tightly kept inactive as a “proenzyme” form and/or by binding to inhibitory proteins (named inhibitors of apoptosis ) in the cytosol. During apoptosis, caspases are activated by cleavage that changes the proenzyme form to the enzyme form and by dissociation of the inhibitors of apoptosis. One of the major apoptotic pathways is derived from mitochondria, which contributes to the activation of caspases in the cytosol. This mechanism is initiated by the release of proapoptotic factors from mitochondria, including cyt c, Smac/Diablo, and HtrA2/Omi. Cyt c released to the cytosol activates caspases through its binding to apoptotic protease activating factor 1 (Apaf-1). Smac/Diablo and HtrA2/Omi bind to the inhibitory proteins to remove their inhibitory effect from caspases.
The apoptotic DNases, including apoptosis inducing factor (AIF) and endonuclease G (Endo G), are also released from mitochondria, in addition to the aforementioned proapoptotic factors. The release of these proapoptotic factors is regulated under the tight control of mitochondrial membrane permeability. This increase in the mitochondrial membrane permeability is mediated via at least three distinct mechanisms: (1) physical rupture of the OMM as a result of mitochondrial swelling (usually linked to mPTP opening); (2) a modification of the structure of voltage-dependent anion channel (VDAC) through interaction with proapoptotic proteins such as the Bcl-2 protein family; and (3) formation of a new pore as a consequence of oligomerization and membrane insertion of proapoptotic proteins, including the Bcl-2 family of proteins. The first pathway involves ΔΨm depolarization and swelling of the matrix space, followed by loss of OMM integrity and rupture, spilling proapoptotic proteins into the cytosol. In contrast, the other two pathways occur without ΔΨm depolarization.
The MCU gene (known as CCDC109A ) encodes a 40-kDa protein containing the mitochondrial target sequence (MTS) at the N-terminus. , The characteristics of MCU are summarized as follows: (1) MCU proteins form ruthenium red-sensitive Ca 2+ channel pore by oligomerization; (2) knockdown of MCU dramatically reduces mitochondrial Ca 2+ uptake; and (3) the changes in MCU expression levels do not affect basal ΔΨm , oxygen consumption, ATP synthesis, or mitochondrial morphology. Significant roles of MCU on mitochondrial Ca 2+ accumulations were confirmed in various cell lines and in primary cells, including cardiomyocytes. , The MCU topology was investigated by several groups, and it seems likely that both the N- and C-termini of MCU face the mitochondrial matrix and a short motif of amino acids for the pore-forming region faces the intermembrane space. Advances in structural biology techniques, including cryogenic electron microscopy (cryo-EM) and nuclear magnetic resonance (NMR), have allowed for an investigation of the architecture of the core region of MCU. MCUs oligomerize and likely form a pentameric homo-oligomer, tetramer, or dimer-of-dimer architecture that generates its pore.
Rizzuto’s group reported a novel protein called MCUb, which served as an endogenous dominant-negative pore-forming subunit of mtCUC. MCUb, which is encoded by a related gene of CCDC109A called CCDC109B , is a 33-kDa protein that has two predicted transmembrane domain (TMD), shares 50% similarity to the MCU with the key amino acid substitutions in the pore-forming region critical for its Ca 2+ conductance, and shows different expression profiles from the MCU in various tissues. The function of the MCUb was characterized by several groups and summarized as follows: (1) MCU and MCUb bind each other and form hetero-oligomers, altering the mtCUC pore structure ; (2) incorporation of MCUb into mtCUC decreases the association of MICU1/2 to alter channel gating ; and thus (3) MCUb inhibits mtCUC activity. Importantly, the ratio of MCU and MCUb mRNA expression varies in different tissues, which might correlate to the size of mitochondrial Ca 2 + -selective currents recorded from mitoplasts isolated from a variety of mouse tissues. For instance, cardiac and skeletal muscle are organs that possess a high copy number of MCU mRNA, , , but the MCUb expression in the heart is much higher compared with in the skeletal muscle. Accordingly, the MCU current measured in whole mitoplast from hearts was 30 times smaller than that from skeletal muscle. These observations suggest that the ratio of MCU and MCUb expression may be one of the mechanisms that characterize different mitochondrial Ca 2+ uptake profiles in different tissue types, including the heart.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here