Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The origins of intact fetal circulating trophoblast cells and nucleated red blood cells in maternal plasma are reviewed, and their pitfalls and promises for noninvasive prenatal testing are discussed.
The discovery of cell-free fetal DNA (cffDNA) in maternal circulation and how it has allowed the development of an excellent noninvasive screening test for common aneuploidies and sex chromosomal abnormalities in both the high-risk and general populations is discussed.
The clinical performance of cffDNA is reviewed, including sensitivity, specificity, positive predictive value and false-positive and false-negative rates.
The different techniques using cffDNA to detect fetal aneuploidies and factors affecting the fetal DNA fraction are discussed.
The challenges of using cffDNA testing to identify fetal copy number variants are presented, including the depth of sequencing. Clinical use of cffDNA to identify subchromosomal abnormalities is currently not recommended by major societies.
Prenatal screening to identify pregnancies at risk for fetal cytogenetic abnormalities has been practiced for almost half a century, although the approach has constantly evolved with the goal of increasing the detection of fetal aneuploidies while simultaneously decreasing unnecessary false positives. Fifty years ago, advanced maternal age was the sole risk factor used in population-based screening, and identification of pregnancies with Down syndrome was the primary goal. At that time, the detection rate was approximately 30% with about 5% of the population at risk because of advanced maternal age. By the 1990s, second trimester multiple serum markers (human chorionic gonadotrophin (hCG), alpha-fetoprotein (AFP), unconjugated estriol (uE 3 )) were added to modify the maternal age risk, which improved detection of trisomies 21, 18 and 13 to approximately 80% for the same 5% screen positive rate. Subsequently, the addition of nuchal translucency (NT) measurement to first trimester serum biochemical analytes (pregnancy-associated placental protein A (PAPP-A) and hCG) brought these detection rates up to 92% to 95%. (These screening methods are reviewed extensively in Chapter 19 .)
Despite the excellent screening performance of indirect biomarkers, acquisition of fetal tissue or DNA would allow complete genetic analysis of a fetus. Presently, this is only possible using invasive diagnostic procedures such as chorionic villus sampling or amniocentesis. Despite multiple attempts over the past 20 years to retrieve and isolate fetal cells from the maternal circulation or cervix, this has not proven clinically viable. A major paradigm shift has recently occurred in which the analysis of fetal cell-free DNA (cfDNA) circulating in the maternal circulation has been demonstrated to detect more than 99% of Down syndrome pregnancies with less than a 0.1% false-positive rate (FPR). This screening method has been rapidly introduced into clinical care and is presently recommended by major societies worldwide for use in pregnancies at high risk for common whole-chromosome aneuploidies (trisomies 21, 18 and 13). This includes populations such as women of advanced maternal age, those with positive biochemical and NT screening, those with a previous offspring having a common trisomy, a known parental balanced Robertsonian translocation increasing the risk for an unbalanced offspring involving chromosomes 21 or 13 or fetuses with ultrasonographic findings indicative of an increased risk for aneuploidy ( Table 21.1 ). Its use in lower risk populations is now being considered because data are progressively showing that its technical performance is equally reliable. Further work in the development of noninvasive prenatal diagnostic testing using cfDNA is ongoing with the ultimate goal of detecting other cytogenetic abnormalities similar to those attainable with direct fetal testing.
| Major Society | Recommendations |
|---|---|
| ACOG/SMFM (2015) | NIPS is currently only recommended as a screening option for women at increased risk for fetal aneuploidy. |
| SOGC (2013) | NIPS should be an option available to women at increased risk as an alternative to amniocentesis. |
| ISPD (2015) | NIPS may be considered in women classified as high risk based on serum and ultrasound screening, contingently to women considered high or intermediate risk after conventional screening or all pregnant women. |
| RCOG (2014) | No specific recommendations (ROCG Scientific Impact Paper No. 15) |
| ACMG (2016) | Inform all pregnant women that NIPS is the most sensitive screening option for traditionally screened aneuploidies. (ACMG Policy Statement) |
| ESHG/ASHG (2015) | No specific recommendations (ASHG/ESHG Joint Statement) |
The passage of fetal material, including intact cells, across the so-called ‘placental’ barrier into the maternal circulation has become a focus of intense research over the past 20 years because retrieval of a small number of such cells would provide a complete copy of the fetal genome amenable to analysis. This has the potential to be a true diagnostic test using improved molecular technologies such as chromosomal microarray and sequencing.
The first report of fetal cells in the maternal circulation occurred in 1893 by a German pathologist Georg Schmorl when he described the presence of trophoblast cells in maternal lungs in patients dying from eclampsia. Not until 1969 was the presence of fetal cells in the circulation of pregnant women with normal pregnancies confirmed with the identification in maternal blood of fetal lymphocytes containing Y chromosome material. It was subsequently recognised using fluorescence in situ hybridisation (FISH) and polymerase chain reaction (PCR) that fetomaternal cellular trafficking occurs in all gestations.
Multiple studies have shown the paucity of fetal cells in maternal blood. Their prevalence is estimated at 1 fetal to 10 4 to 10 8 maternal mononuclear cells. To quantify the prevalence of fetal cells, Emad and colleagues used automated scanning of microscopic slides to determine the concentration of fetal cells in maternal blood. With this approach, the number of fetal cells identified per 1 millilitre of maternal blood ranged from 3 to 6 in normal pregnancies and up to 13 to 21 cells in pregnancies affected with Down syndrome. Because of the rarity of these cells, most research on noninvasive prenatal diagnosis using fetal cells has focused on cell separation techniques to both enrich the population of fetal cells and to deplete contaminating maternal cells with the goal of finding a sufficient population of the ‘ideal’ fetal target cells for testing.
Four types of circulating fetal cells have been described: trophoblasts, fetal nucleated red blood cells (fNRBCs), leukocytes and undifferentiated stem cells and progenitors. However, the latter two types persist in the maternal circulation up to 27 years postpartum, making them unusable for prenatal diagnosis of a current pregnancy. Prenatal diagnostic research has thus focused on trophoblasts and fNRBCs which are cleared from the maternal blood rapidly after delivery.
By far the biggest challenge with using these rare and fragile cells for noninvasive testing remains their isolation from contaminating maternal cells without damage. The primary approaches have used gradient separation to increase the relative concentration of the cells of interest followed by their isolation through unique cell surface or cytoplasmic markers ( Fig. 21.1 ).
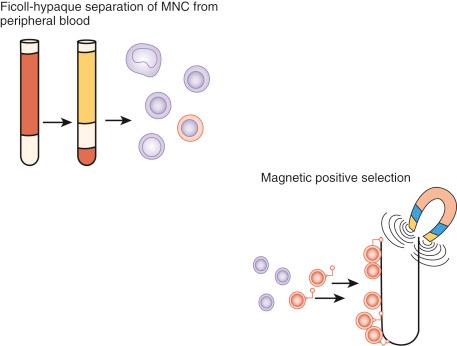
Trophoblasts were the first fetal cell type to be detected in the maternal circulation but present challenges to their use for noninvasive prenatal diagnosis. First, few highly specific antibodies are available for isolation and enrichment. Second, these multinucleated cells are not easily amenable to standard cytogenetic techniques, such as FISH, but require molecular techniques such as chromosomal microarray. Third, because they are placental in origin, there is a 1% incidence of confined placental mosaicism similar to that seen with chorionic villus sampling. Despite these difficulties, contemporary studies continue to show their potential value.
Paterlini-Brechot’s group reported the genetic diagnosis of 63 fetuses at risk for either cystic fibrosis (CF) or spinal muscular atrophy (SMA) using circulating trophoblast cells. Recovery of trophoblast cells was performed by the initial isolation of epithelial tumour/trophoblastic cells (ISET) by size using a calibrated filter. Cells were subsequently laser dissected off the filter and genotyped using maternal and paternal short tandem repeats (STRs) to confirm those that were trophoblast. Of the cells greater than 15 μM in size selected by filtering, half were confirmed as fetal. These cells then underwent specific diagnostic testing. Approximately, 1.5 cells per millilitre of maternal blood were present, and approximately 5 to 10 cells were analysed per case. All fetuses affected by CF or SMA were correctly diagnosed and confirmed by chorionic villus sampling (CVS).
Further evidence of the potential value of trophoblast recovery was presented by Hatt and colleagues using specific markers for cells most likely belonging to the endovascular subgroup of extravillous trophoblast (EVTs). Expression of the endothelial/vascular marker of these originally ectodermal cells is the result of adaptation to their vascular environment. Fetal cells were isolated from maternal blood (gestational age, 11–13weeks) using magnetic cell sorting enrichment (with a novel antibody combination for the endothelial/vascular markers CD105 and CD141). Trophoblast cells were demarcated using a cocktail of cytokeratin antibodies. In 85% of the samples, it was possible to obtain X and Y signals by FISH for gender determination with a 91% specificity. Concordance to fetal sex of 100% was possible if three or more fetal cells could be found in a sample.
Recently, Beaudet and his group developed methods for the detection of chromosomal and subchromosomal abnormalities in fetal trophoblast cells in maternal circulation using array comparative genomic hybridisation (CGH), next-generation sequencing (NGS), or both. They first separated nucleated cells from maternal blood collected at 10 to 16 weeks’ gestation by density fractionation and then immunostained them to identify cytokeratin-positive and CD45-negative trophoblasts. Array CGH, NGS, or both was used for analysis after whole-genome amplification and genotyping and identified normal fetuses and fetuses affected with Klinefelter syndrome; trisomies 21, 18 and 13; and chromosome 15 deletion syndrome.
The presence of trophoblast cells in cervical mucus has also been reported, first by Shettles in 1971 using quinacrine mustard fluorescent staining for identification. The cells were readily identified by their morphology and unique immunohistochemistry using specific antibodies. However, since then, identification of fetal cells in cervical mucus has been inconsistent with detection rates between 50% and 60%.
More recently, Bolnick and colleagues published results on 56 women between 5 and 20 weeks of gestation in whom cervical specimens were collected using a cytobrush and human leukocyte antigen G trophoblast-specific antibodies to identify cells. The average number of cells recovered per patient was 746 ± 59 across the gestational ages. There was minimal maternal cell contamination, and 95% to 100% of the cells isolated expressed confirmatory fetal-specific parameters. Nonetheless, to date, technical challenges have prevented this technology from developing into a clinical tool. Although some groups are still exploring this approach, none has consistently demonstrated success.
Fetal nucleated red blood cells have been detected in the maternal circulation and have a relative short half-life, making them specific to the current pregnancy. This has led to their evaluation as a target for prenatal diagnosis. The advantages of this cell are that they can be identified through a marker profile which is characteristic for erythroid precursor cells and which varies from other blood cell subpopulations. For example, fNRBCs express the transferrin receptor (CD71) on their cell surfaces and do not express CD45 as do maternal leukocytes.
Stringent isolation criteria are required to isolate and identify the fetal cells which are present at a concentration of 1 in 10 5 to 10 7 maternal nucleated cells. After initial staining with cell-specific markers, enrichment of fetal cells most often uses immunomagnetic or flow cytometric cell separation techniques, either alone or in combination. Other approaches attempted include separation by centrifugation using a Ficoll gradient, filtration on a chip, lateral displacement and magnetophoresis, lectin-binding, dielectrophoresis, micromanipulation, laser microdissection and pressure catapulting. This relatively large assortment of approaches shows the lack of consensus on the single best approach for isolation.
Confirmation of the fetal origin of the cells is required after enrichment because maternal immature nucleated RBCs are present and can express markers similar to those on fetal cells such as the transferrin receptors. Immune labelling with embryonic or fetal globin antibodies followed by selected cell dissection or capture is presently the most frequently used approach.
Initially, cytogenetic techniques such as FISH were used for cytogenetic evaluation but proved difficult because of the cellular disruption that occurred during separation and isolation. More recently, molecular approaches which only require a small amount of fetal DNA have proven more efficient.
The National Institute of Child Health and Human Development Fetal Cell Isolation Study (NIFTY) is the largest to date to evaluate fNRBC-based techniques for noninvasive prenatal diagnosis. Results showed that the sensitivity and specificity of the cell-based methods were not satisfactory for aneuploidy detection. In the NIFTY trial, the authors used various potentially unique NRBC cell surface identifiers and both magnetic-activated cell sorting (MACS) and fluorescent-activated (FACS) approaches. The results of this trial, which included 2744 samples, showed an aneuploidy detection rate by FISH of trisomies 13, 18 and 21 and the sex chromosomes of 74.4% with an FPR of 0.6% to 4.1%. This study demonstrated the limitations of NRBC identification, separation and analysis using approaches available at the time. Isolation of fetal nucleated cells for prenatal diagnosis continues to be a costly, labour-intensive and time-consuming process and will remain so until reliable automated approaches become more readily available.
In the past decade, various groups have developed new antibodies to fNRBCs and have used advanced single-cell analytic techniques to continue to explore the possibility of retrieving and analysing fetal cells. One example is the life science biotechnology company RareCyte, which has developed an innovative method using sequential density fractionation to recover intact fetal cells. A two-step centrifugation method takes advantage of the nucleated cells’ density to concentrate and separate cells of interest away from plasma and maternal RBCs. Multiplex imaging using proprietary software then detects and ranks potential cells of interest by analysing individual cell’s morphology and biomarker expression profile. After discovery and characterisation, cells of interest are individually retrieved for additional downstream analysis.
KellBenX focuses on a proprietary monoclonal antibody (4 B9) found uniquely on fNRBCs and their precursors and not on surface antigens of maternal RBCs. When the antibody is bound to an epitope expressed by fNRBCs, fetal cells can be isolated and can undergo full analysis by sequencing, PCR, FISH, microarray or immunohistochemistry.
Although these advancements are intriguing, they have not yet proven useful for clinical analysis because the complexity of the approaches and the need to validate that the retrieved cells are exclusively fetal make it difficult to scale for routine screening.
Circulating cell-free nucleic acids in plasma and serum have been described in other fields of medicine, such as oncology and trauma, as novel biomarkers for various diseases. Mandel and Metais were the first to report the presence of extracellular nucleic acids in the circulation in 1948. In 1997, real-time PCR targeting SRY, a single-copy Y-chromosome-specific sequence, was used to demonstrate and quantify the amount of fetal DNA in pregnant women carrying male fetuses. This discovery of cell-free fetal DNA (cffDNA) in the maternal plasma opened a new perspective in prenatal diagnosis.
The main advantage of cffDNA is that it is up to 1000-fold more prevalent than the DNA obtained from the limited number of circulating fetal cells. It also diminishes rapidly after birth with a half-life estimated to be 16.3 minutes. cffDNA levels are undetectable by 2 hours postpartum, making it specific to the current pregnancy.
Initially, cffDNA fragments were examined by conventional or real-time quantitative PCR to identify fetal DNA sequences that were completely absent from the maternal genome (e.g. the Y chromosome from a male fetus or the fetal RhD gene absent from Rhesus-negative pregnant women). Since then, the clinical utility and success of cffDNA analysis has continued to expand with the detection of fetal aneuploidies, sex chromosomal abnormalities and more recently microdeletions and microduplications.
Many cells go through a life cycle, which ends in programmed cell death called ‘apoptosis’. During this process, DNA gets cleaved into short fragments of 150 to 200 base pairs that are released into the blood as ‘cell-free DNA’ (cfDNA). During pregnancy, DNA fragments arise from cytotrophoblast cells of the placenta through a constant turnover of villous trophoblasts and are released into the maternal circulation, resulting in maternal plasma containing a mixture of both maternal and cffDNA. The sum of the fetal DNA fragments in the maternal circulation represents the entire fetal genome.
The term cell-free ‘fetal’ DNA is misleading when applied to that found in the maternal circulation because the DNA emanates predominately from the placenta. It is therefore not surprising that cffDNA is elevated in a number of conditions associated with placental pathology such as preeclampsia, preterm labour, placenta previa, hyperemesis gravidarum and fetal intrauterine growth restriction.
The amount of cffDNA in the maternal plasma represents approximately 3% to 10% of the total and occasionally may reach 30%, depending on multiple factors. cffDNA can be reliably detected after 10 weeks of gestation; however, reports have shown detection as early as 5 to 7 weeks.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here