Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Familiarity with the basics of nucleic acid biochemistry and biology is required to understand molecular diagnostic testing.
The chemical stability of double-stranded DNA stands in contrast to the lability of single-stranded RNA.
Base pairing of nucleic acids is dictated by energetically favorable rules and forms the basis for DNA replication, RNA transcription, and hybridization assays.
The chemical similarity of nucleic acid molecules, regardless of their source, means that methods of extraction, storage, and handling are often similar.
Enzymes that synthesize and modify nucleic acids (e.g., polymerases, transcriptases, nucleases, ligases) may be harnessed as tools for molecular biology and molecular diagnostics.
Nucleic acid analyses include electrophoresis, hybridization assays, amplification techniques, sequencing, and polymorphism detection. Complete assays or diagnostic tests often combine several of these techniques.
Molecular diagnostics is now part of the mainstream of laboratory diagnostics.
Nucleic acids are the critical molecules of life. Deoxyribonucleic acid (DNA) resides in the nucleus of eukaryotic cells and maintains all the information necessary for maintenance of the organism and for transfer of the information to successive generations. Ribonucleic acid (RNA) carries information from DNA to the cytoplasm of a cell and directs synthesis of the proteins necessary for the function of the organism. The normal state of health depends on the stability of DNA and on accurate DNA duplication and translation into protein. Modern cell biology seeks to determine the basic mechanisms of cell structure and function. Studies are increasingly focused at the level of the gene, the protein-coding units of DNA. Therefore, diagnostic methods are also being directed toward nucleic acid evaluation. The goal of this chapter is to provide the conceptual framework for diagnostic applications of nucleic acid analyses.
Nucleic acid biochemistry is central to modern cell biology and dictates many aspects of diagnostic applications. Many of the enzymes associated with in vivo nucleic acid synthesis, degradation, and repair have become basic laboratory tools for manipulation and analysis of DNA and RNA. Cellular mechanisms that operate to direct and control DNA replication, transcription, and translation address the basic biology of the cell in health and disease. Diagnosis, therapy, and research are all increasingly directed at a molecular level of cellular function. This chapter provides an overview of those aspects of nucleic acid biochemistry and biology that are required to understand the current diagnostic applications of molecular biology. Further details are available from textbooks on cell biology (e.g., ).
DNA is a long, double-stranded polymeric molecule (dsDNA) that exists predominantly in the form of a right-handed double helix. Each single-stranded DNA molecule (ssDNA) is composed of a small number of building blocks. The backbone of the ssDNA polymer is the sugar deoxyribose connected by phosphate groups ( Fig. 68.1A ). Phosphodiester bonds between the 3′ carbon of one sugar ring and the 5′ carbon of the next give the backbone its invariant structure and its 5′ to 3′ directionality. Linked to the 1′ carbon of each sugar is one of four possible bases: thymine (T) and cytosine (C), which are pyrimidines, and adenine (A) and guanine (G), which are purines. The bases can occur in any sequence order and, thus, form the variable portion of ssDNA. The building blocks of the single-stranded polymer are the four deoxyribonucleotide triphosphates (dTTP, dCTP, dATP, dGTP), each consisting of a sugar molecule, a triphosphate group, and one base. During DNA synthesis, nucleotides are first stripped of two phosphate groups and then enzymatically linked together by phosphodiester bonds to form a chain.
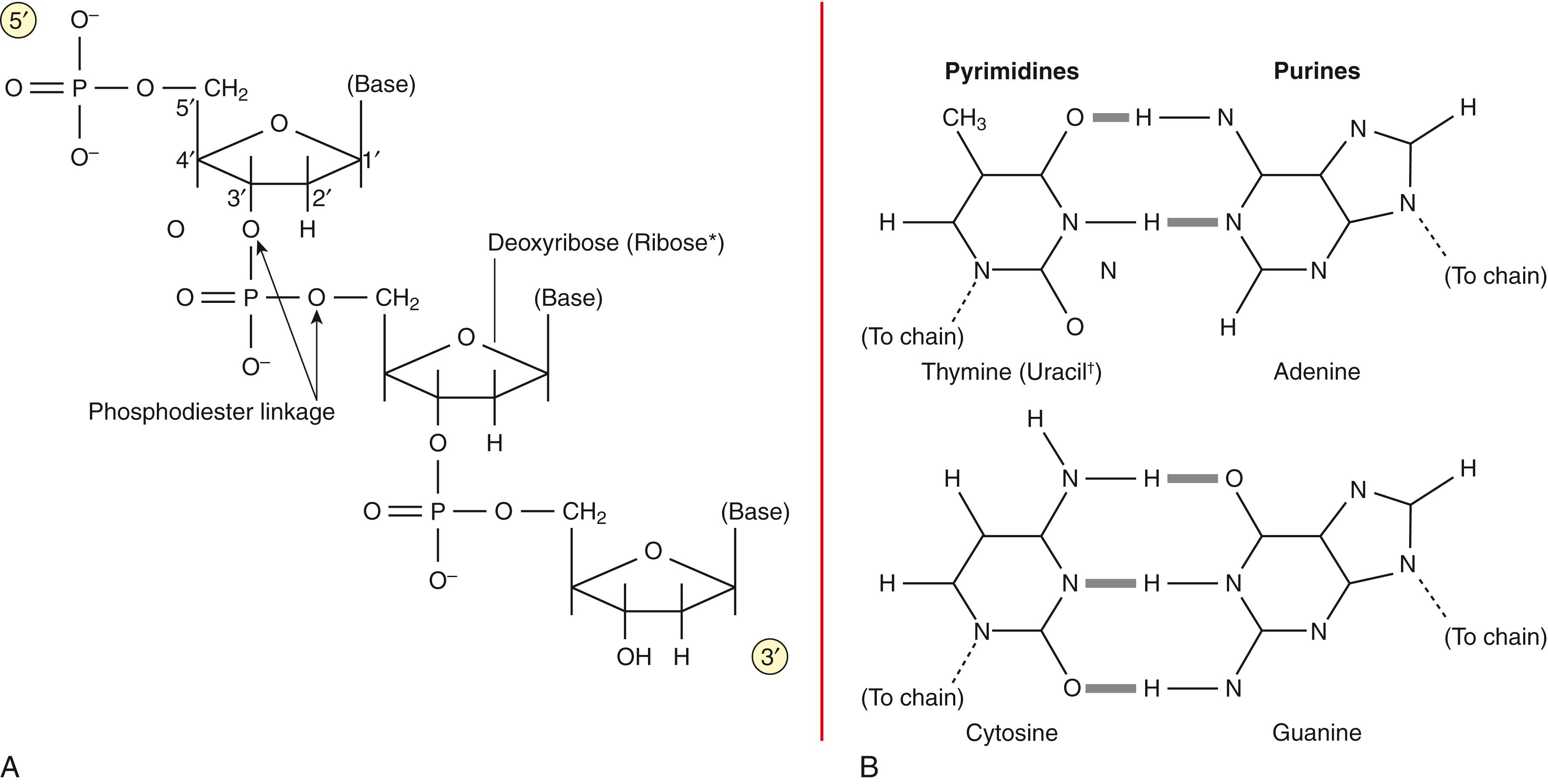
DNA is an extraordinarily stable molecule, losing its normal conformational structure only at extremes of heat, pH, or in the presence of destabilizing agents. The double-stranded helix is the most energetically favorable state for DNA—an examination of the components of DNA explains this fact. Both sugar and phosphate groups are hydrophilic, forming stable hydrogen bonds with surrounding water molecules in solution. Bases, however, are hydrophobic and are not soluble in water at neutral pH. A stable molecule of DNA must ensure that the bases do not contact water. This is possible when two antiparallel ssDNA polymers (one running in the 3′ to 5′ direction, and the other 5′ to 3′) twist around the same axis. This arrangement allows planar hydrogen bonds to form between adenine and thymine, and between guanine and cytosine (see Fig. 68.1B ). As long as the two chains have base sequences in complementary order, the strands of the helix have a ladder-like structure with rungs (base pairs [bp]) of consistent size. The flexibility of the carbon-oxygen linkages in the phosphodiester bond allows the ladder to twist, forming a regular helix such that the planar base pairs are stacked on top of each other, leaving no room for water molecules in between. The polymeric series of base pair hydrogen bonds holds the ssDNA chains tightly together, and the helical conformation protects the base pairs from water, exposing only the hydrophilic backbones.
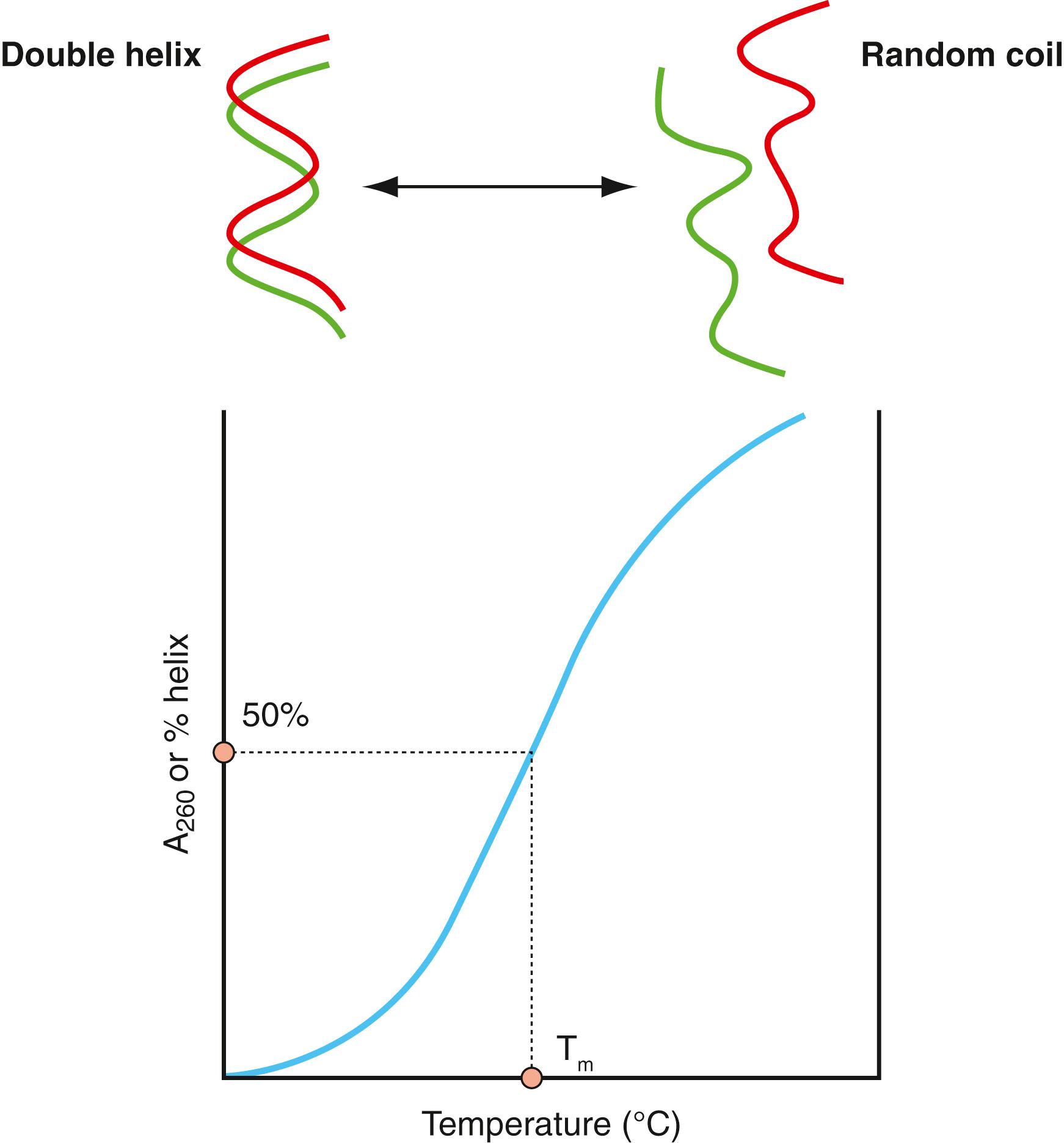
Helical dsDNA is stable over a pH range of approximately 4 to 9. Solutions with pH outside these limits have the capacity to disrupt the base pair bonds and cause the DNA helix to denature or unwind into two separate, random coils. Extreme heat and hydrogen bond disrupters, such as formamide, also have the same effect. This helix-to-coil transition can be followed spectrophotometrically at A 260 ( Fig. 68.2 ). The bases absorb ultraviolet (UV) light maximally at this wavelength but at a lower molar absorptivity in dsDNA; absorptivity increases 20% to 30% when dsDNA is converted to ssDNA. Because temperature is often used to effect this transition, the process has been referred to as melting , and the temperature at which 50% of dsDNA is converted to ssDNA is called the melting point or T m of the DNA. The T m of DNA molecules depends on the relative G-C versus A-T base pair content, because the three hydrogen bonds of G-C base pairs require more energy to disrupt than the two hydrogen bonds of A-T base pairs. Lowering the temperature can reverse the melting process, and the two complementary strands reform the original helix if the base pairs re-form in the correct linear conformation.
The length of a fully extended eukaryotic DNA molecule would be about 3 m per genome, much longer than the cell itself. Undegraded purified DNA forms a stringy viscous solution, reflecting the extreme length of genomic DNA ( Fig. 68.3 ). In vivo, however, DNA is organized into highly compacted, regular units called chromosomes . A chromosome is composed of its DNA strand wound around DNA-associated proteins in a highly structured fashion (chromatin). The chromosome structure involves several hierarchical levels of chromatin packing, from a “beads-on-a-string” nucleosome (146 nucleotide pairs wound around a histone core) to highly condensed loops, each containing about 100,000 bp of DNA. Each human cell nucleus contains two sets of 23 chromosomes of characteristic length and unique base pair sequence. Together, these chromosomes constitute the human genome . The complete sequence of the 3 billion chemical base pairs that make up the human genome was finished in 2001. (The Human Genome Project is discussed in Chapter 80 .)
RNA differs from DNA in chemical composition, structure, and function ( Table 68.1 ). In RNA, the sugar is ribose, containing a hydroxyl group at the 2′ position, and thymine is replaced by the methylated uracil (U). RNA exists predominantly as a single-stranded molecule and in much shorter lengths than DNA. The structure of RNA is more irregular, owing to the single-stranded nature of the molecule, but it may contain some helical sections and hairpin loops. However, RNA molecules of the same base sequence will form the same three-dimensional structure as a result of adopting the most energetically favorable conformation. These consistent three-dimensional structures are a requirement for RNA function. RNA is much less stable than DNA, due not only to the single-stranded structure but also to its susceptibility to alkaline hydrolysis via the 2′ hydroxyl group of the ribose moiety. Ubiquitous RNA-specific enzymes also rapidly degrade RNA.
| Feature | DNA | RNA |
|---|---|---|
| Sugar | Deoxyribose | Ribose |
| Base pairs | Thymine–adenine | Uracil–adenine |
| Cytosine–guanine | Cytosine–guanine | |
| 3D structure | Double-stranded | Single-stranded |
| DNA duplex helix | Variable, determined by base sequence (see text) | |
| Stability | Stable | Subject to base hydrolysis |
| Degraded by DNase | Degraded by RNase | |
| Function | Maintains genetic information in nucleus | Carries genetic information to cytoplasm |
DNA must be duplicated prior to cell division so that daughter cells retain an exact copy of the genetic information contained in parent chromosomes. RNA must be synthesized by all functioning cells to direct the synthesis of necessary proteins. DNA must be degraded during repair of damaged segments, and RNA is continually degraded and resynthesized. Enzymes that operate directly on nucleic acids affect these and other functions. Table 68.2 lists major categories of nucleic acid–specific enzymes and their in vivo functions. In vitro, purified enzymes have become laboratory tools for the molecular biologist, allowing genetic engineering and facilitating many nucleic acid assays for research and clinical use.
| Enzyme | In vivo function |
|---|---|
| Polymerases DNA polymerases RNA polymerases |
Polymerases join DNA or RNA nucleotides together to form a single-stranded daughter molecule, using a stretch of single-stranded parent molecule as a template. These enzymes perform syntheses according to base pair rules and proceed in the 5′ to 3′ direction. |
| Reverse transcriptase | Mostly of viral origin, reverse transcriptase catalyzes the synthesis of DNA from either an RNA or DNA template. |
| DNA ligases | Joins DNA fragments formed by discontinuous synthesis in DNA replication or by DNA repair pathways. |
| Nucleases DNases, RNases |
Nucleases “digest” nucleic acid molecules by breaking phosphodiester bonds. |
| Endonucleases Exonucleases |
Endonucleases digest nucleic acids from the middle of the molecule, whereas exonucleases begin at a free end and may require a 3′ or 5′ end. Nucleases may have single-stranded, double-stranded, DNA, or RNA specificity. Some polymerases also have nuclease activity. |
| Restriction endonucleases | Bacterial endonucleases that recognize specific short DNA base pair sequences and cleave the DNA molecule only at the recognition site. |
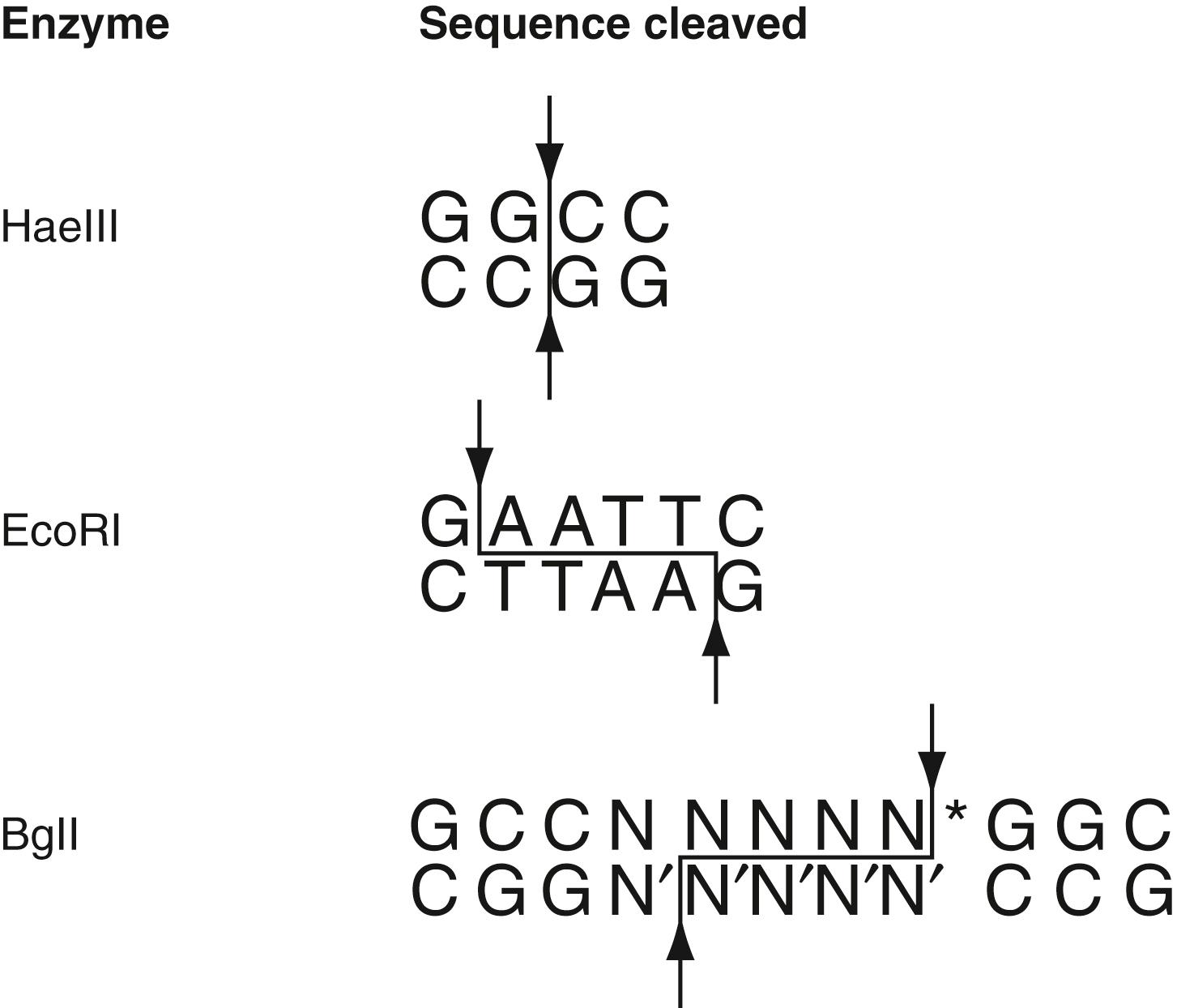
Nucleic acid–specific enzymes include polymerases, which catalyze the formation of phosphodiester bonds during synthesis, and nucleases, which hydrolyze these bonds. RNA-specific nucleases (RNases) are present virtually everywhere. Because of this, it requires much greater care in laboratory practice to work with RNA in vitro than with DNA . Restriction endonucleases are a special category of nucleases found only in bacteria, where they function to destroy foreign DNA. The recognition sites for restriction enzymes are located anywhere within the DNA molecules, are sequence specific, and can vary approximately 4 to 12 bp in length. At the recognition sequence or nearby restriction endonucleases make specific asymmetric or blunt end cuts in DNA molecules ( Fig. 68.4 ). More than 3000 restriction endonucleases have been discovered and categorized according to the structure of their corresponding sequence recognition sites ( ). The in vivo function and in vitro utility of many of these enzymes are discussed in the following sections.
The DNA duplication process, known as semiconservative replication , uses each strand of the parent molecule to direct the synthesis of a daughter strand ( ). Because the base sequence of the parent strand dictates the sequence of the daughter strand, replication is faithful and the replication products consist of two dsDNA molecules composed of one parent strand and one daughter strand each, with exactly the same base pair sequence. Although this is conceptually simple, the process is complicated and involves a number of accessory proteins and enzymes. First, for synthesis to begin, a small single-stranded region must be produced at a point on the genome called an origin . This is not energetically favorable and must be accomplished using proteins—that is, helicases—that unwind and separate the strands of the helix. Next, at each origin, a short RNA primer is synthesized complementary to the single-stranded sequence. DNA polymerase III proceeds with DNA synthesis; later, the RNA primer is excised and replaced with DNA by DNA polymerase I. Chromosomal DNA contains many origins of replication. This process occurs simultaneously across the chromosome. Interestingly, DNA polymerase III is a directional enzyme and can synthesize DNA only in the 5′ to 3′ direction because it requires a free 3′OH end. This means that only one daughter strand, called the leading strand , can be synthesized continuously. The opposite strand, called the lagging strand , is primed by RNA primase that does not require a free 3′OH end. It is synthesized discontinuously in short fragments (Okazaki fragments), each requiring an RNA primer, as the replication fork is opened up. These fragments are then joined together by DNA ligase after each primer is excised. DNA polymerase III is also unique in that it has proofreading and exonuclease activity. If an incorrect nucleotide is added to the growing chain, it is detected and excised by the nuclease portion of the enzyme; the correct nucleotide is then added. This helps explain the extraordinary fidelity of the DNA replication process. Postsynthesis repair mechanisms also contribute to the accuracy of replication (see Mechanisms of DNA Repair section). Finally, telomerase adds DNA repeat sequences to the 3′ end of each chromosome (the telomere) to prevent shortening of the lagging strand with each replication cycle.
Sections of DNA that specify amino acid sequences of proteins are called genes . One gene contains the amino acid sequence code for one protein as well as the DNA sequences necessary for the regulation of the production of that protein. Within the 6 billion nucleotides of the human genome, there are about 20,000 to 25,000 protein-coding genes. Although these coding sequences are of paramount importance to the cell and to the function of the organism as a whole, they actually make up less than 2% of the nucleotides. The vast majority of the human genome is composed of noncoding DNA regions referred to in the past as junk DNA . While much of the function of noncoding DNA remains unknown, diverse roles of biological importance have been identified, such as regulation of gene expression, origins of replication, and coding of RNA (see later section on Gene Regulation Mediated by Small RNA and ).
Protein synthesis begins with the activation of the appropriate gene. A copy of the gene is made from DNA in the form of RNA. Because the RNA copy carries the code from the DNA in the cell nucleus to the cytoplasm where amino acid synthesis takes place, this type of RNA is called messenger RNA (mRNA). mRNA is synthesized from only one strand (coding strand) of the DNA gene; the complementary DNA (cDNA) strand is not used. This is accomplished by a process called transcription . DNA promoter sequences present near the start of the gene to be transcribed promote the ability of RNA polymerases and associated proteins to recognize the nucleotide at which mRNA synthesis initiation begins. Synthesis of mRNA proceeds in much the same fashion as DNA replication, with the ssDNA sequence dictating the mRNA sequence using the same rules of base pair complementarity (uracil base pairs with adenine). When the end of the gene is reached, mRNA synthesis is terminated. Some genes are always expressed, while others are only active in certain physiologic situations. The rate of transcription also varies in different cells (see later section on Transcriptional Control).
Before export to the cytoplasm, the mRNA molecule is modified in several ways ( ). mRNA contains both amino acid coding sequences (exons) and noncoding sequences (introns); introns are excised from the mRNA molecule before protein synthesis. A molecular complex called a spliceosome ( ), which is composed of both low-molecular-weight RNA (including, at its catalytic core, a ribozyme; see ) and protein, recognizes mRNA sequences that identify the boundaries of an intron, joins the flanking exons, and releases the intron. Splicing must be exact, because the addition or subtraction of a single nucleotide at the splice junction would change the 3-nucleotide reading frame in the nucleotide sequences that follow.
Further modifications to the mRNA molecule include the addition of 7-methyl guanosine residues to the 5′ end in a unique 5′-5′ phosphodiester bond. This is called a cap and aids in the binding of the ribosome to the mRNA molecule for initiation of protein synthesis. A poly-A tail, which may be necessary for stability and transport to the cytoplasm, is added to the 3′ end. The polyadenylation locus is specified in part by the sequence AAUAAA, usually found in the 3′ untranslated region of the RNA transcript. At this point, the mRNA molecule is ready to direct the synthesis of its corresponding protein.
Protein synthesis requires translation from the language of nucleotides to that of amino acids. Twenty-one different amino acids are used in protein synthesis; each amino acid is specified by one or more mRNA nucleotide triplets called codons . For example, AAG is the codon for lysine, and UCG is the triplet for serine. Thus, an amino acid coding sequence is read in groups of three nucleotides running in the 5′ to 3′ direction; this is the reading frame of a protein coding sequence. Three specific codons—UAG, UGA, or UAA—do not code for amino acids but instead signal the end of a gene ( stop codons). Because an amino acid can be encoded by more than one codon, the code is referred to as a degenerate code .
Translation from the mRNA nucleotide code to protein is mediated by ribosomes in the cytoplasm of the cell. A ribosome binds to the 5′ end of the mRNA and provides a stable chemical environment for all molecules involved in protein synthesis. Amino acids are linked in the correct sequence by the action of small adaptor RNA molecules called transfer RNAs (tRNAs). Each tRNA molecule contains a region that is complementary to a particular mRNA codon: the anticodon . Linked to one end of the tRNA is the amino acid that corresponds to the complementary mRNA codon of the tRNA. A tRNA with the correct complementary anticodon binds to the first codon in the mRNA sequence, which is always AUG. When another specific tRNA binds to the next codon, ribosomal enzymes catalyze the formation of a peptide bond between the two amino acids linked to the tRNAs, removing the linkage between the first amino acid and its tRNA molecule. The first tRNA is ejected from the ribosome, and a new tRNA binds to the next codon. Protein synthesis proceeds from the N-terminus to the C-terminus. As this process continues, the ribosome moves along the mRNA molecule, completing the synthesis of the amino acid chain. When a stop codon is reached, the ribosome detaches from the mRNA. In reality, several ribosomes can move along the same mRNA molecule, each translating the mRNA code into a new protein molecule.
To allow for cellular differentiation and response to environmental stimuli, there must be mechanisms controlling the repertoire of gene transcription and protein translation. Some of these mechanisms operate at the level of DNA and control the transcription of mRNA ( ). As noted, promoters are important for the initiation of mRNA transcription and are found upstream (toward the 5′ end) at a relatively invariant distance from the beginning of the protein coding sequence. There are several different kinds of promoter consensus sequences (nucleotide sequences that are found in many examples). The most common promoters are rich in adenine and thymine and have been called TATA boxes . Because A-T base pair bonds are weaker than G-C base pair bonds, DNA unwinds more easily at repeat A-T sequences. After transcriptional activation, a local ssDNA region is produced and stabilized by the assembly of a complex of RNA polymerase and general transcription factors ( Fig. 68.5 ). mRNA is synthesized from the local region of ssDNA, and the mRNA is quickly ejected as the DNA returns to its more energetically favorable double-stranded helical state.
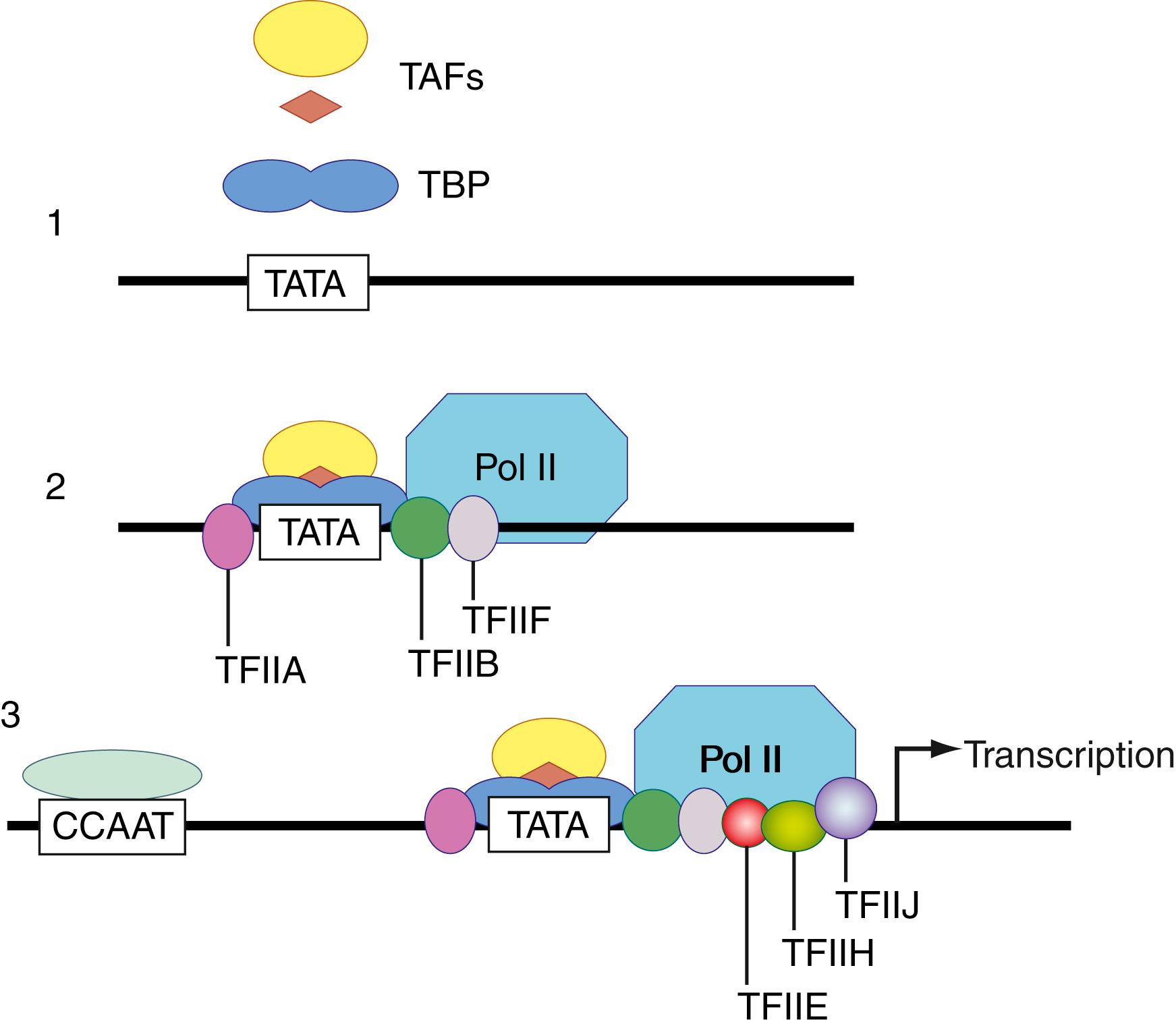
Enhancers are DNA sequences that can augment mRNA transcription and may be found in different locations relative to the gene that they affect. Gene-specific transcription factors are proteins that bind to enhancers and promoters and selectively stimulate or inhibit mRNA transcription ( ). In eukaryotes, enhancers commonly contain multiple binding sites for several transcription factors, so it is the net effect of factors binding to these DNA elements that determines the activation and rate of expression. The availability of transcription factors, in turn, is controlled by cellular events such as phosphorylation or by other proteins such as hormones and growth factors. A network of intracellular and extracellular chemical communications can thus select and control the synthesis of necessary proteins.
Because mRNA is far less stable than DNA, the half-life of mRNA is very short. New mRNA molecules are continually transcribed from DNA. As the cell responds to changes in transcriptional signals, the genes that are transcribed into mRNA can be quickly changed, resulting in the immediate synthesis of new proteins. Thus, the cell has the ability to rapidly adjust its protein output in response to its environment.
An additional mechanism of gene regulation by another class of RNAs—small RNAs—is recognized as important. Small RNAs are short, noncoding ribonucleotides that function as posttranscriptional regulators of gene expression. First discovered in the 1990s, they were found in cells of many eukaryotic organisms, including mammals ( ). The small RNAs are divided into several different subgroups, most notably small interfering RNA (siRNA) and microRNA (miRNA). Recently, next-generation sequencing has identified large numbers of noncoding RNAs of greater length (>200 nt). The function of these long noncoding RNAs (lncRNA) is an active area of research ( ).
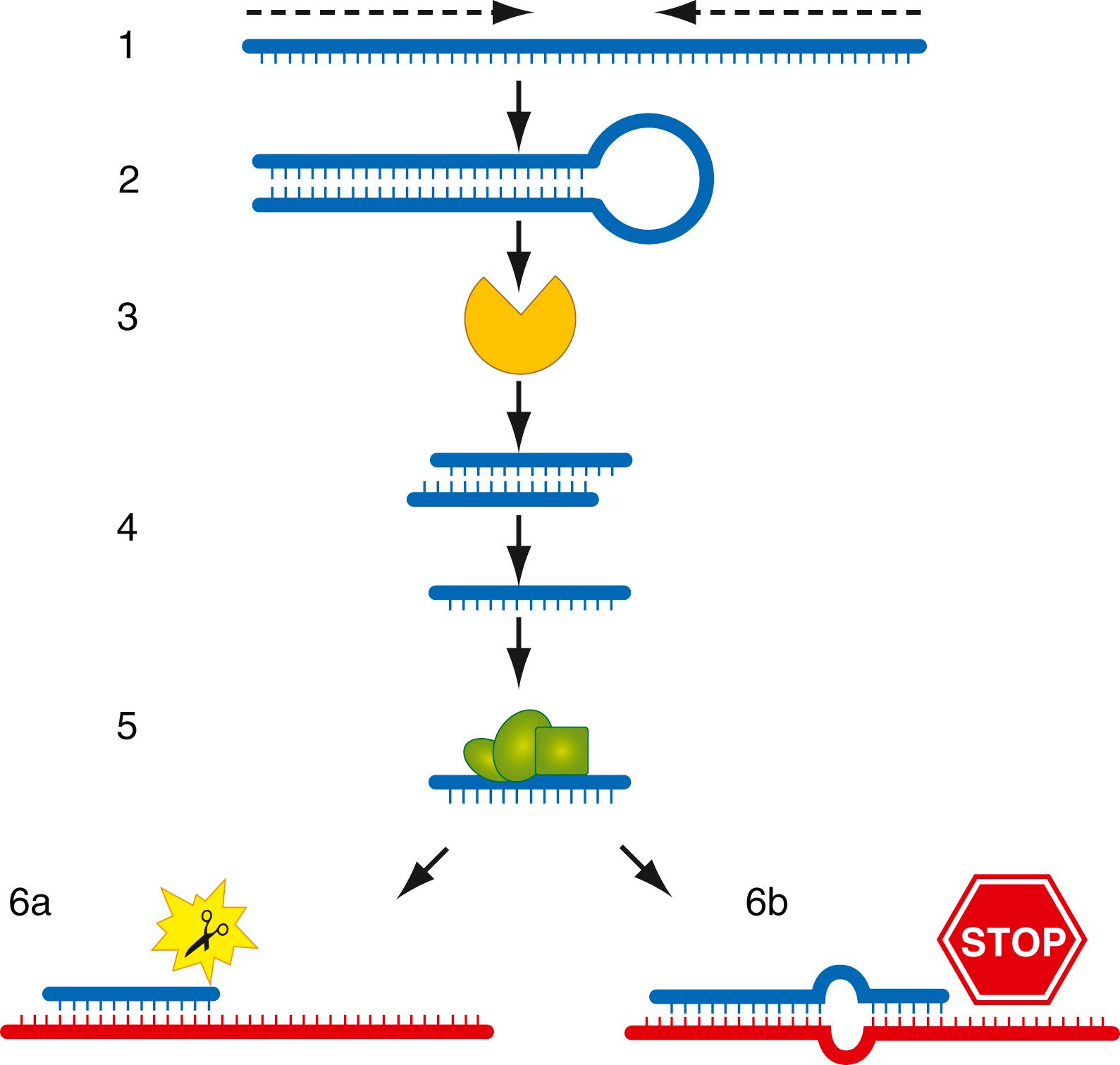
The DNA sequences for siRNAs and miRNAs are often located in noncoding regions between genes. They generally have their own promoters, and transcription is mediated by polymerase II. The immediate product is an miRNA precursor that forms a double-stranded hairpin structure ( Fig. 68.6, 6a ). An enzyme called Dicer processes it to short RNA duplex molecules and further into single-stranded RNAs 22 to 26 nucleotides in length. The small RNA strand attaches to an RNA-induced silencing complex (RISC), which mediates binding to complementary sequences of mRNAs. siRNAs pair perfectly with the target mRNA and trigger a series of molecular mechanisms that lead to its degradation. This process is also called RNA interference ( RNAi ). miRNAs bind imperfectly to their target mRNA, causing conformational changes, bulging, and subsequent blockage of protein translation at the ribosome. Small RNAs are abundant in eukaryotic cells (several hundred); it is thought that each one can target multiple mRNAs either as siRNA or miRNA. They are often tissue specific and have been implicated in the regulation of all major cellular processes such as proliferation, differentiation, metabolism, and cell death. siRNAs have also been recognized as a new RNA-based immune system against viruses that is based on silencing through the RNAi mechanism. Furthermore, changes in miRNA expression patterns can be found in many malignancies, and specific miRNAs were found abundantly in certain tumors ( ).
miRNA research has advanced within a decade from one publication to thousands of publications describing the role of miRNAs in gene regulation. miRNA expression profiling has been recently evaluated as a reliable diagnostic biomarker for differentiating between normal and tumor specimens ( ). It has shown to be deregulated in multiple cancers in human and mouse models, and has proved to play a critical role in the development and progression of the tumor. Most of the miRNAs are differentially expressed, whereas some discriminate totally between normal and tumorigenic samples. The miRNA expression enhances (oncogenic miRNA) or reduces (tumor suppressor) as the tumor progresses and has been found to be associated with drug resistance. This discovery of miRNA a decade ago, as a diagnostic and prognostic marker, has now led to miRNA-based targeted therapy in vitro and may selectively predict better treatment outcome for patients with cancer. In addition, classification of an unknown tumor may be possible by the alteration of tumor-specific miRNA. The nomenclature for assigning names to novel miRNAs for publication in peer-reviewed journals is done by miRBase, which is the central repository for miRNA sequence information. It has an online database with all published miRNA sequences with links to the primary literature and to other secondary databases. Although some miRNAs are tumor specific, miR-21 has proved to be the global oncogenic miRNA in many solid tumors. The miRNAs with oncogenic potential include miR-155, miR-17-92, and miR-21 but it is not limited to these alone. The level of expression of miR-155 is upregulated in various carcinomas. However, they are specifically significant in pancreatic cancers, for which they have prognostic relevance.
Several synthetic small molecule inhibitors of miRNAs, such as chemically modified antisense oligonucleotides called “antagomiRs,” are currently in use in vitro, targeting against specific oncogenic miRNAs. These antagomiRs silence the overexpressed oncogenic miRNAs in cancers by blocking their function. In animal experiments, antagomiRs against miR-16, miR-122, miR-192, and miR-194 were found to be efficacious in reducing the levels of miRNA in the liver, lung, kidney, and ovaries. Re-expression of tumor-suppressor miRNA, such as let-7, is another proposed miRNA therapeutic strategy to upregulate tumor-suppressor miRNA by exogenously transfecting let-7 that led to the inhibition of growth in vitro and in vivo. These characteristics of miRNA suggest their potential role as novel biomarkers for diagnostic, prognostic, and therapeutic targets ( Fig. 68.6, 6b ).
Epigenetics refers to changes in gene expression that are not dependent on changes in DNA sequences. In most instances, this involves a change in the chromatin structure that facilitates or blocks gene transcription. This is a dynamic process involving enzymes that chemically modify DNA or histone proteins. A methyl group added to the fifth carbon of cytosine results in 5-methyl cytosine. This generally occurs on cytosines located next to guanines (CpG) that are often repeated in islands or patches located in the promoter region. DNA methylation tends to result in condensed chromatin and reduced transcription. DNA methyltransferases are the enzymes responsible for DNA methylation. Chromatin proteins may be acetylated, deacetylated, or methylated. There is evidence that small regulatory RNA may directly interact with a variety of epigenetic mechanisms ( ). Epigenetic changes are recognized to be important in tissue-specific gene transcription; alterations may be factors in cancers, aging, and stress response. The relationship of the microbiome has further been shown to promote epigenetic changes associated with disease and to modify gene expression patterns in a state of disturbed immunity (e.g., inflammatory bowel disease). To this end, poor dietary choices are encoded into the human gut and genetic make-up, which could be transferred to offspring in an epigenetic influential manner ( ; ; ).
Errors in DNA replication and damage to DNA during the normal cellular lifetime must be minimized to preserve the health of the entire organism. Several mechanisms operate to maintain the normal DNA sequence. First are the error-avoidance mechanisms that operate during DNA replication. The DNA polymerase that synthesizes new DNA polymers selects each successive nucleotide monomer based on its complementarity to the next nucleotide in the template strand. Fidelity at this level is high, and most errors in synthesis are avoided at this stage. Nevertheless, an occasional base may be incorrectly added to the growing strand. To adjust for this, the proofreading activity of the polymerase can recognize the error, remove the incorrect base, and proceed again with synthesis. Together, the error avoidance mechanisms reduce base pair mismatches to approximately 1 in 10 million ( ). This represents a 100,000-fold increase in efficiency compared with the error rate of 1 in 160 bases for in vitro solid-phase oligonucleotide synthesis.
Despite the remarkable error avoidance in DNA replication, occasional mistakes do occur. In addition, DNA can be damaged by normal biochemical reactions and by nonphysiologic agents such as UV light and environmental carcinogens. Several repair mechanisms mend damaged DNA ( ) ( Table 68.3 ).
| Repair Type | Function |
|---|---|
| Direct repair | Repairs certain types of DNA damage in a single-step reaction. |
| Mismatch repair | Checks for errors made when DNA is replicated. Any mispaired bases in the daughter strand are removed and replaced with the correct match. |
| Base excision repair | Repairs small, nonhelix-deforming adducts such as those produced by methylation, oxidation, reduction, or base fragmentation by ionizing radiation. |
| Nucleotide excision repair | Removes bulky DNA adducts such as thymine dimers and certain photoproducts as well as chemical adducts and cross-links. |
| Double-strand break repair | Repairs double-strand breaks that result from physiologic processes or from ionizing radiation and oxidative insults. |
Direct repair mechanisms repair lesions in a single-step reaction. For example, O 6 -methylguanine DNA methyltransferase repairs alkylation lesions by transferring the alkyl group from the lesion to the active site of the enzyme. Other direct repair mechanisms characterized in Escherichia coli may also exist in human cells ( ).
Mismatch repair (MMR) functions immediately after DNA replication to replace mismatched bases with the correct ones ( ). Several MMR proteins recognize the error in the newly synthesized daughter strand and excise a region that includes the mismatch. DNA polymerase III and ligase restore the correct sequence and integrity of the daughter strand. The importance of this mechanism in stabilizing the genome is evidenced by recent studies associating Lynch syndrome with defects in MMR proteins.
Base excision repair targets small, nonhelix-deforming adducts such as those produced by methylation, oxidation, reduction, or base fragmentation by ionizing radiation. Base excision leaves three or four nucleotide sequence gaps that are then filled with the correct nucleotides; nicks are sealed with ligase. Larger, bulky adducts or dimers that distort the DNA helix may be caused by UV radiation, carcinogens, and therapeutic drugs, among other agents. Such damage is removed by the nucleotide excision repair (NER) pathway, which uses an enzyme system composed of many proteins to excise a single-stranded oligonucleotide containing the lesion ( ). The gap is then filled in by DNA polymerase and ligated. There are two NER pathways: one in which lesions that block transcription are rapidly removed (transcription-coupled repair; ) and a global pathway that repairs bulk DNA, including the nontranscribed strand of active genes. Some of the possible consequences of the loss of NER activity are exemplified by the disease xeroderma pigmentosum (XP), which is caused by mutations in NER. XP results in extreme sensitivity to sunlight, with skin cancers occurring at an early age ( ).
Double-strand breaks that occur rarely or are produced by ionizing radiation and oxidative damage present significant repair problems. If unresolved, replication and transcription of involved sequences will be blocked. To maintain local and overall genomic integrity, double-stranded breaks are repaired by nonhomologous or allelic recombinational repair mechanisms. One of the genes associated with hereditary breast-ovarian cancer syndrome, BRCA1 , encodes a protein involved in repairing double-strand breaks in DNA and illustrates the importance of this process.
It is now clear that DNA repair plays a central role in the life of the cell. Recent evidence also indicates that several primary repair proteins also function in transcription and in regulation of the cell cycle. Thus, the processes involving DNA appear to be highly integrated and are increasingly studied as a whole and in relation to human disease.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here