Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Over the past decade, the human microbiome has been discovered to be a major factor in human health and disease. The human microbiome consists of complex communities of bacteria and microorganisms that inhabit the human body including viruses, fungi, protozoa, and archaea. These microbial communities have the ability to occupy specific niches within the body as a result of a lifetime of coevolutionary strategies that have been shaped by host and environmental factors resulting in diverse populations that differ depending on location on and within the body. The human microbiome is a complex and dynamic system in which the bacteria, for example, are dependent on their host, the neighboring communities of microorganisms, and the specific environmental conditions needed to grow and thrive. Due to the complexity of the microbiota’s ecosystem, standard culture methods have failed to identify the majority of the bacteria, viruses, and fungi that make up the microbiome. The rise of next-generation sequencing has allowed for the characterization of the bacteria (and in some cases fungi and viruses) that make up the microbiome. As a result, changes to the microbiome are now able to be interrogated such that they can be associated with major disease states such as critical illness and sepsis and correlations to outcome determined. For example, the combination of the traumatic injury and the associated surgical interventions and hospital care following the injury likely have a profound impact on a patient’s microbiome. Next-generation technology with sequencing and metabolomics can now allow for dynamic tracking of the microbiome along the continuum of care and its role in the occurrence, course, and outcome of critical illness and injury determined. In this chapter, we will give a brief introduction to the microbiome and highlight how the microbiome affects the trauma patient, predominantly focusing on the gut microbiome as this is the field with the largest body of evidence related to trauma patients.
The term “microbiota” can be used to describe the whole body’s microbial population by family, genus, or species level and can refer to specific populations that occupy an area or body region of interest. In general, the term “microbiota” refers to the normal inhabitants of a bacterial community, whereas the term “pathobiota” indicates the presence of pathogens among the community. The term “microbiome” refers to not only the members of a given microbial community, but its structure in terms of the relationship of members to one another and its function in terms of the genes that are expressed and the metabolites they produce. Given the large body of studies and evidence demonstrating the impact of the gut microbiome on human health, the majority of studies in this chapter will focus on the gut and its bacteria, recognizing that the microbiome includes archaea, viruses, and fungi. Regardless of the particular microbiome being studied, the two main features of the microbiota that are commonly reported are structure and function ( Fig. 1 ). The structure (microbial community members’ relative kinship to one another) is determined by the microbial species present and is determined through the use of next-generation sequencing, typically 16S rRNA sequencing or shotgun metagenomics. Bacterial species possess a 16S ribosomal subunit whose unique sequence variations can be used to classify the bacteria down to the species level. Therefore, microbiota composition is reported as a relative abundance at the phylum, class, order, family, genus, or species level. Microbial community structure is also reported as a measure of bacterial diversity, typically alpha or beta diversity. Alpha diversity, or intrapopulation diversity, takes into consideration the species richness or the number of different species present within a single environment or sample and the evenness across the population of the species represented. Alpha diversity is frequently reported as a numeric score that indicates how many different types of bacterial species are present. Beta diversity, or interpopulation diversity, is a comparison of diversity across environments or samples. Beta diversity is measured using several different indices that account for differences in taxonomic abundance profiles and is generally depicted in the form of a principal component analysis plot. Principal component analysis is a graphical representation of the variables, such as level of diversity, that explain the variation that exists between samples. Samples are individually plotted on an ordination plot, and those with similarities cluster together at nearby positions on the plot.
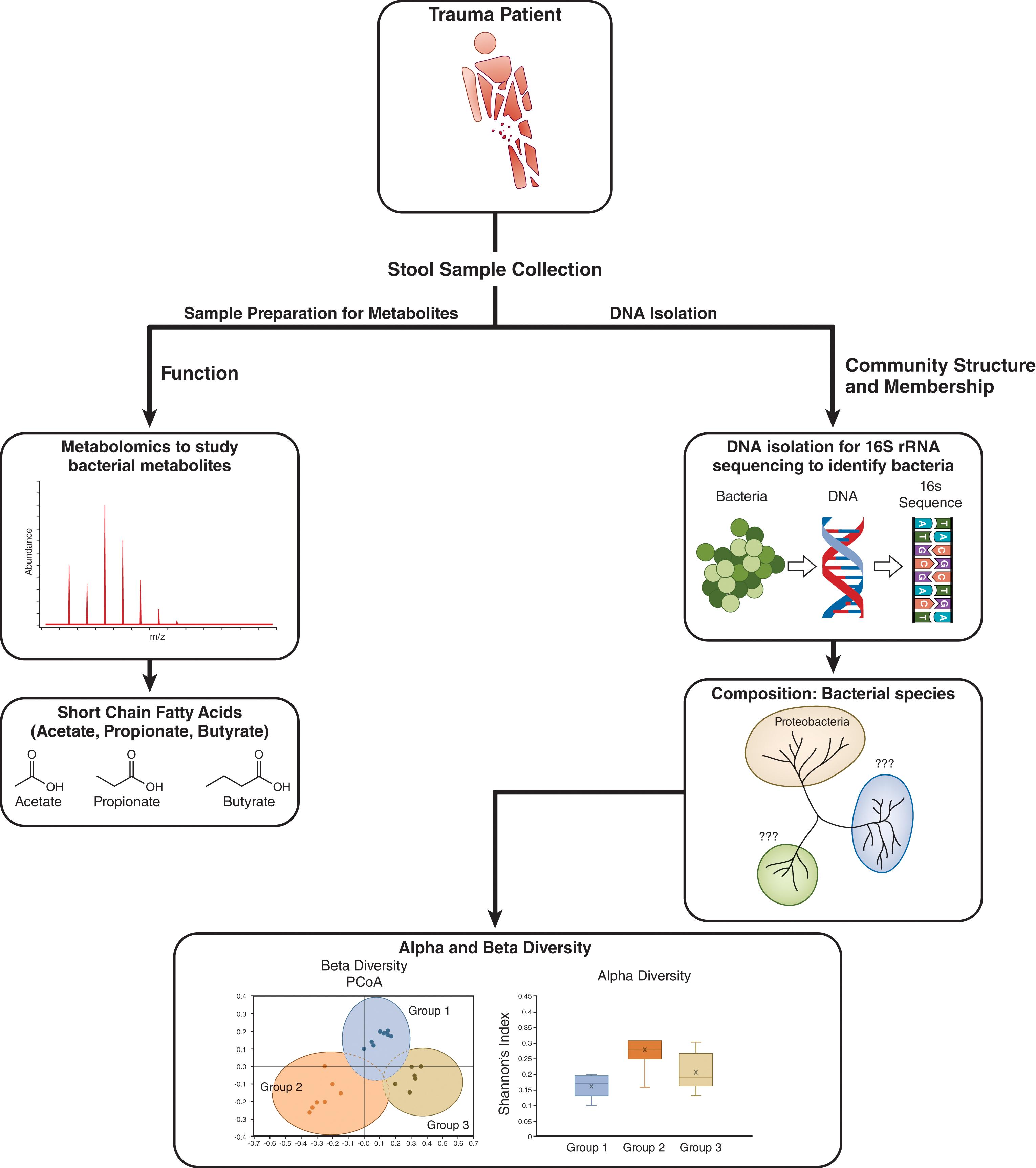
The measures of composition via speciation and diversity give a broad-brush stroke of the bacteria that are present; however, they are limited in their ability to advance our understanding of the actual functional output of bacteria within a given population or sample. A more complete understanding of microbiota function can be important given the dynamic interaction between bacterial communities and the host. The most widely studied area of microbiota function is bacterial metabolism, particularly the metabolism that results in the production of short-chain fatty acids (SCFAs) (acetate, butyrate, and propionate) from fermentable carbohydrate. There is also a wide interest in the ability of the microbiota to conjugate bile acids, which also have important interactions among bacteria. In understanding the production of these metabolites, the function of the microbiota can be inferred from genomic data, determined by gene expression via metatranscriptomics (gene expressions across all microbes within a given community or sample), or directly measured by mass spectrometry. The functional assessment of the microbiota is a continually emerging field and is providing a greater level of detail of how the microbiome impacts the host.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here