Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The large number of test results generated by clinical laboratories has led to challenges in data management and analytics. Because of the potential diagnostic value of examining these results in aggregate, it is important to utilize emerging tools for the analysis of high-dimensional data. Machine learning uses a variety of computational algorithms to analyze complex datasets and make robust predictions.
This chapter discusses the varied definitions of big data and their application to laboratory medicine. It also presents workflows, concepts, common algorithms, infrastructure, and applications related to the use of machine learning in the clinical laboratory. The chapter is a more technical and extensive version of one previously authored on these topics. Because each biomarker measured in a patient can be plotted as a single number, the collection of biomarkers measured in this patient can be represented in a high-dimensional space. Unsupervised learning methods are used to find patterns in this high-dimensional space, and supervised learning methods use known outcomes from a set of subjects to develop a model to predict the outcome in a new, unknown subject. A variety of different algorithms, each with different advantages and disadvantages, has been used for these machine learning tasks. Implementing machine learning in the laboratory requires not only understanding the basic algorithmic concepts, but also deploying an appropriate computational infrastructure. Machine learning has been successfully deployed in laboratory medicine settings using a variety of underlying datasets, including traditional laboratory values, next-generation sequencing data, and images.
Laboratory data associated with an individual patient can be considered a point in a high-dimensional space
Developing machine learning models involves a process of data collection, training, and testing
Machine learning methods can be used to address both classification and regression problems
Unsupervised learning methods look for inherent structure within high-dimensional data
Supervised learning methods train a classifier based on data associated with a known target outcome
The modern clinical laboratory routinely generates large amounts of patient data, and this rise in volume has been driven by both increases in the number of processed samples and by technological developments in high-throughput analyzers and in highly multiplexed assays. In addition to data sources within the laboratory, increasing amounts of data from other diagnostic modalities (such as noninvasive imaging) and clinical phenotype data have become readily accessible through the electronic medical record (EMR). As a result, laboratorians have more comprehensive information than ever before on a larger group of cases, and the increase in data has led to an increasing reliance on computational tools to understand and interpret the data.
The traditional laboratory has made great progress in handling the operational aspects of high-throughput laboratory medicine, including monitoring patient samples and identities, running validated assays and providing guidance on their use, monitoring for factors that may change the validity of results, and delivering results to the treating clinician in a timely fashion. However, the increasing volume of data often means that there is insufficient real-time analysis and diagnostic integration of these complex results. Further, despite the increasing amount of data potentially available for any given patient from the clinical laboratory, the task of integrating multivariate results often falls to the treating clinician.
The advent of computational approaches, such as machine learning, provides an opportunity for the clinical laboratory to improve the nature of the diagnostic information provided for patient care. In this chapter, we will discuss the nature of large data sources within the clinical laboratory and provide an overview of the tools of machine learning along with their use.
It is important to consider diagnostic implications in order to understand the potential value of integrating multiple laboratory results (or, more generally, multiple biomarkers) into a single result.
A single biomarker that completely separates two diagnostic populations (say, presence vs. absence of disease) can achieve perfect sensitivity and specificity ( Fig. 13.1 A). However, once the values of this biomarker overlap between diagnostic categories ( Fig. 13.1 B), the sensitivity and specificity drop, and the area under the curve (AUC) decreases to less than 1. If two single biomarkers have similar overlaps between categories ( Fig. 13.1 C), then it would appear that neither one adds significantly to the diagnostic value compared with the other. However, it may be the case that when considered in two dimensions, these biomarkers are individually similar in performance, can provide an aggregate diagnosis that fully separates presence versus absence of disease ( Fig. 13.1 D). Further, even biomarkers that appear completely uninformative on their own ( Fig. 13.1 E) may be informative when assessed in a multidimensional way ( Fig. 13.1 F).
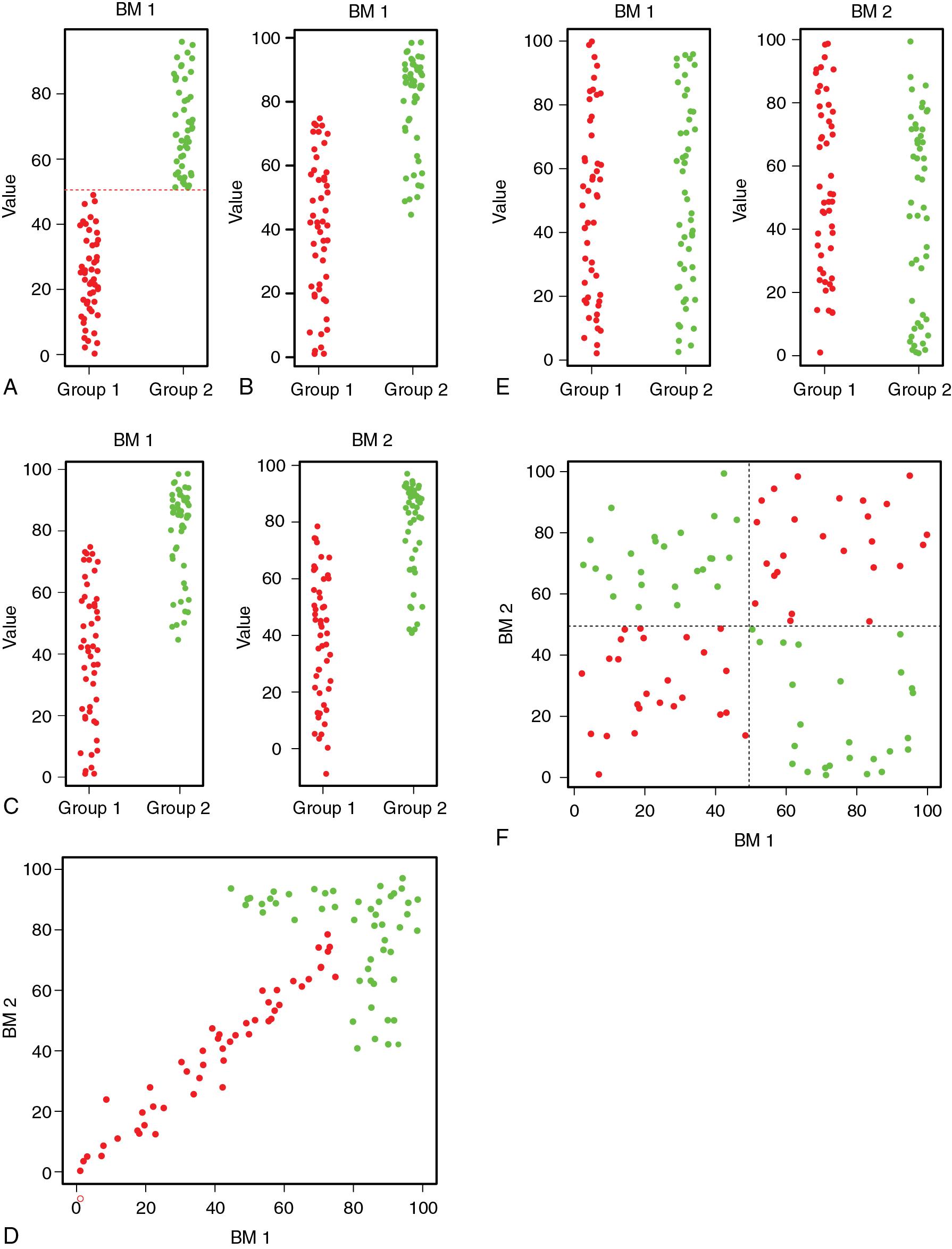
This multidimensional view may be extended with increasing numbers of biomarkers. As previously shown, a single biomarker may be graphed in one dimension, and two biomarkers can be graphed in two dimensions ( x - and y -axis). More generally, any number of biomarkers can be represented in a high-dimensional space, where the number of dimensions is equal to the number of biomarkers. This is adequate for visualization when the number of dimensions is three or fewer, but for higher numbers of dimensions it is a challenge to understand the relationship between cases that are represented this way. In a subsequent section, we will discuss computational techniques such as dimensional reduction and clustering that can address this problem. For the moment, it is important to understand the reason that large numbers of biomarkers imply a high-dimensional space and to recognize that the lack of human intuition about high-dimensional data leads to a need for computer-based tools.
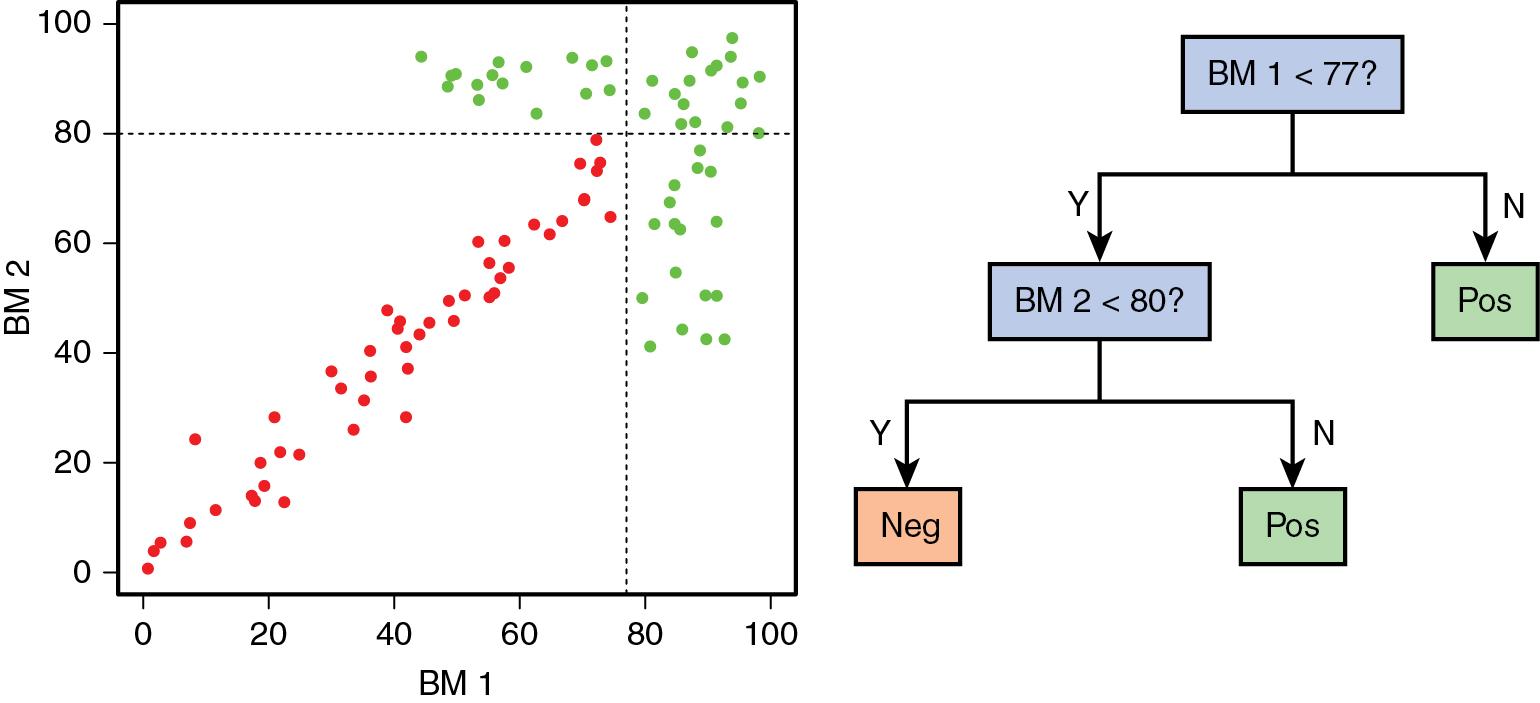
Because of the complex nature of high-dimensional data, it is often necessary to utilize various computational algorithms to understand the diagnostic information. Machine learning refers to the processes that are used to take a set of training cases (typically including the known outcome that is to be predicted), create a computational classifier, and use that classifier to accurately predict the outcome of a new case or set of cases that were previously unknown (note that throughout this chapter we will use the term “cases”; this could refer to patient “samples,” and it is typical in machine learning to refer to a case as an “observation”). As a simple example, the two-dimensional data that we previously described ( Fig. 13.1 C and D) can be encoded using a decision tree ( Fig. 13.2 ), and a new case can accurately be classified. In this instance, machine learning (a term that is sometimes interchangeable with artificial intelligence) takes the observed training data from Fig. 13.1 D, creates the appropriate decision tree, and applies it to classify new cases. Later in the chapter, we will discuss a variety of different machine learning algorithms, along with their various strengths and weaknesses.
There are two basic types of problems addressed by machine learning: classification and regression. Classification problems take the high-dimensional input data set and use it to predict one of a discrete set of outputs. Typically, this is a binary decision such as disease versus no disease, although it is equally possible to have multiple classes. Regression, in contrast, takes the set of input variables and predicts a continuous output. For example, a set of input laboratory values that predicted a patient’s tumor size or length of hospital stay would both use regression strategies. The relationship between regression and classification will be further discussed later in the chapter.
Discussions of machine learning and its application often occur in conjunction with references to big data . At its simplest level, big data simply refers to the large datasets, such as medical datasets, that we described in the introduction and will further elucidate in subsequent sections. However, there are subtle differences in the way that the term big data has been used, and these can be broken into at least three categories: a technology-driven definition, a property-driven definition, and an analysis-driven definition.
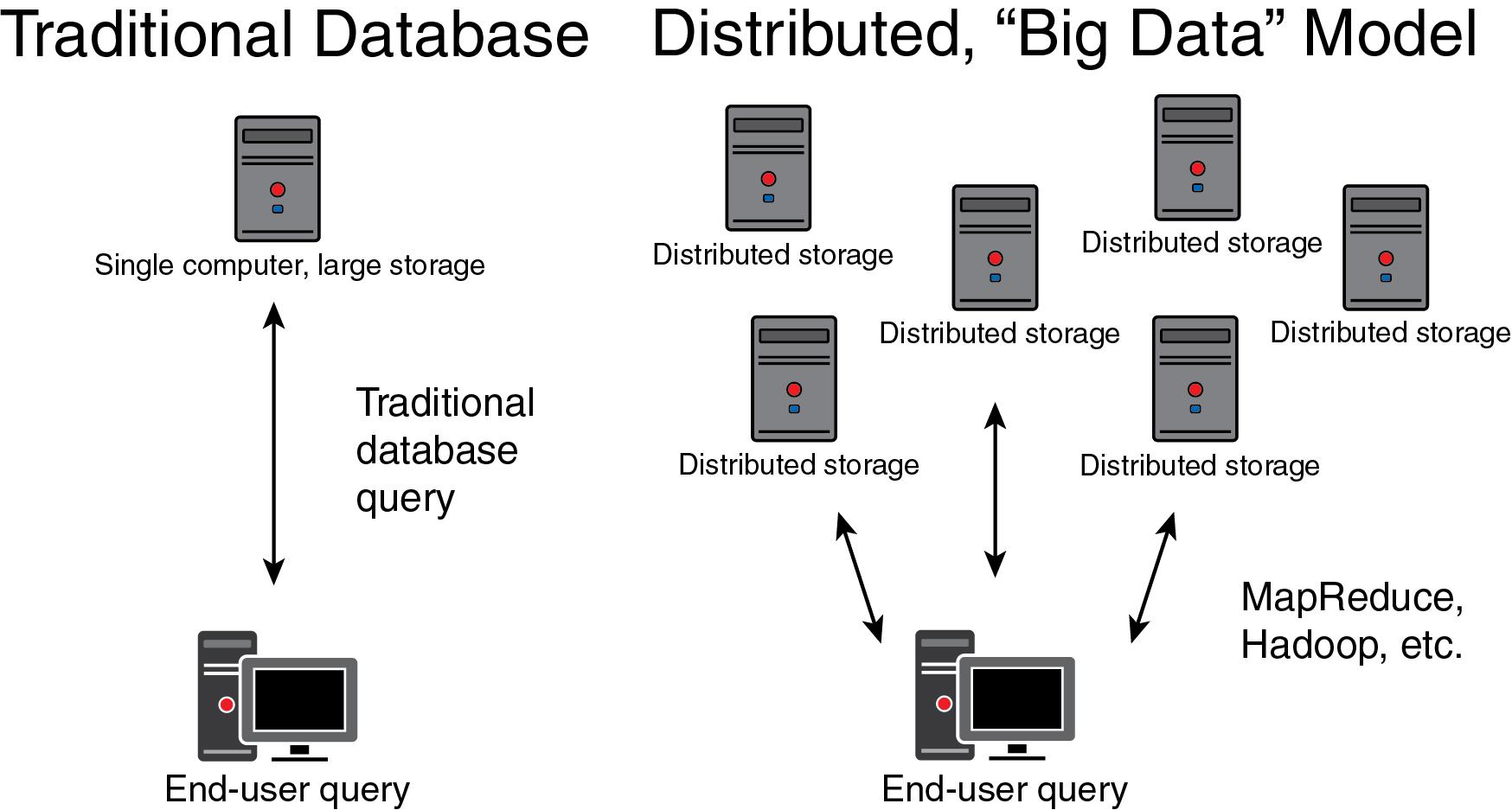
The technology-driven definition refers to the challenges with having a dataset that is too large to reside on a single computer system or a dataset that is too complex for a traditional relational database management system. Big data requires distribution over a larger, networked area ( Fig. 13.3 ). Traditional datasets contain structured data that are easily modeled using a relational format where data is organized into rows and columns, making up one or more tables. Relational databases are manipulated using a standard programming language, SQL (Structured Query Language). In contrast, big data may contain a variety of data types, including those that are unstructured or semi-structured and, therefore not amenable to tabular database structures. This poses fundamental difficulties, compared with traditional data architectures, since any query or computation with the data must coordinate resources that are spread over a larger domain than a single server or storage device and must be able to do so for relational and nonrelational data models. These distributed jobs require parallel processing to increase efficiency, given the number of computational processes typically required of big data . Several software tools (MapReduce, Hadoop, etc.) were specifically designed to overcome the challenges raised by these sorts of distributed processes. , Similarly, nonrelational data structures such as NoSQL have become a critical component of big data architectures because they are well-suited for high throughput use of unstructured and semi-structured data types commonly encountered in big data applications. NewSQL databases have also gained popularity as a new type of relational database structure with the scalable properties of NoSQL. When the technology-driven definition of big data is being used, it is typically in the context of these or similar software tools and data architectures that solve the particular technical challenges imposed by big data . It is certainly true that large health system databases can outstrip traditional storage and computing paradigms, and in this sense the technology-driven definition is relevant. However, these issues are now typically transparent to the end-user, as they are handled by enterprise IT groups, and thus the clinical laboratorian does not ordinarily have to consider the details of the technical solutions that have been developed.
The property-driven definition of big data refers to a number of inherent characteristics of data that are being acquired in modern commercial and other settings. These properties are often summarized by the 4 (or 5) “Vs”: volume, velocity, variety, and veracity (with some commentators adding “value”). , Volume refers to the increasingly large amounts of data that are being produced; in the health care context, the amount of laboratory and clinical data now available is notable. Velocity refers to the rate at which information is being recorded; in a major academic medical center, it is not unusual to generate over 15 million tests per year in a core laboratory alone. Variety refers to the different modalities of data; the combination of numeric laboratory data, textual interpretive data, imaging data from pathology and radiology, and other physiologic measurements contribute to the variety of data within the modern EMR. Veracity refers to the potentially unreliable nature of the quantities of information, often with the assumption that volume, velocity, and variety can help to overcome the limitations posed by issues with veracity. While the clinical laboratory has focused on high-quality data streams, it remains possible that errors have crept into the medical record, and this possibility should be accounted for when assessing the overall patient picture. The property-based definition of big data provides important insights into the nature and quality of data gathered at high scale, and it is important for laboratorians to assess the extent to which their data fit this description.
Finally, the analysis-driven definition of big data uses the term to refer to high-dimensional datasets that are not amenable to human interpretation in the absence of computational tools for efficient data visualization and machine learning. This definition appears to unite the more formal, property-driven definition, which has characterized much discussion in the internet-driven commercial sector, with a recognition of challenges imposed by smaller high-dimensional datasets. These laboratory datasets may be extracted from the laboratory information system (LIS), or they may be genomic, proteomic, and metabolomic datasets which report large number of analytes from a single patient. Interestingly, the analysis-driven definition emphasizes the common set of computational tools that are required in both cases, whether a metabolomics interpretive problem or a traditional big data set that exhibits the 4 Vs. Whether or not the clinical laboratorian finds the property-driven definition directly applicable, the tools of machine learning are critical to the ability to make intelligent use of the data.
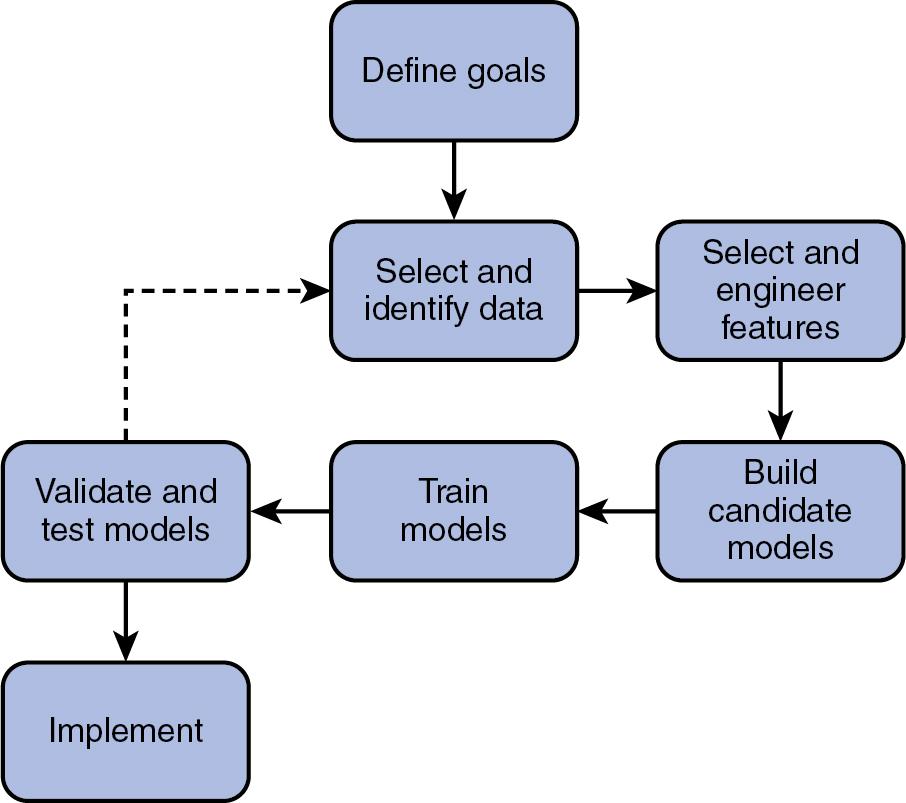
An overview of the machine learning process is shown in Fig. 13.4 . Machine learning approaches start with defining the goals for the model and framing the relevant question as a machine learning problem that will be used to generate a model that is a functional representation of the available data with some amount of error. A suitable source of data must be identified for use in the model. Typical sources within the laboratory include records from the LIS; measurement systems with large numbers of potential data elements, such as mass spectrometry; other high-dimensional data typically associated with the “-omics”; and digital images. Exploratory data analysis and data preparation steps are conducted to help identify or engineer potentially informative predictors (or features) for the candidate models. A portion of the available data needs to be designated as a “training set” that will be used to shape the model parameters as it learns to associate input data with a correct output. There are a large number of actual machine learning algorithms that can be utilized, and each one has slightly different parameters and performance for certain applications. The remaining data may be used as a “validation set” used to select the best machine learning algorithm and associated set of parameters for the problem. Model training and validation results may necessitate reiterating through the process to find a seemingly useful model. Once the best performing model and its parameters are selected, the model must be tested, ideally on an independent “test set” of data, which may be held out from the original data set. In the best case, test data can be independently collected from a variety of settings in order to determine whether the model is able to generalize its predictive performance in new situations. Implementation of a machine learning model requires consideration of end-user needs and available infrastructure. We will address each of these steps in subsequent sections of this chapter.
The increased volume and variety of available data are among the major factors that have driven interest and successful application of machine learning methods in various industries outside of laboratory medicine. Within the laboratory, data have also become more plentiful both at a single time point (with increased numbers of conventional tests or newer, highly multiplexed testing formats) and longitudinally as patients are monitored over time. In turn, we are experiencing growing enthusiasm for using machine learning methods to aid in medical and operational decision making in clinical laboratories. Typical clinical laboratory data for machine learning will be discussed based on their source.
One of the most abundant data sources for machine learning in laboratory medicine is the LIS. LIS databases contain millions of records that are coded and available as highly structured and standardized data elements. Increasingly, organizations are integrating laboratory data with clinical features and outcome data and are aggregating large amounts of this medical data across health systems, creating data repositories ideal for use with machine learning. LIS data repositories also contain information that may not be transmitted to electronic health records for patient care, but are valuable features for predictive analytics in laboratory operations. Examples include documentation of physician notifications of critical results and time stamps for intermediate steps in laboratory processes (in addition to collection, order, receipt, and result times).
LIS data are often obtained through reports that are exported as formatted text (typically comma-separated values; .csv files) or in spreadsheet formats (e.g. .xls). The columns of this format reflect different variables, such as patient name, medical record number, test name, test result, units, upper and lower reference limits, flags, and others. Every row lists a different result, and the format can be easily imported into a number of data analytics packages for analysis and machine learning. A similar form of data can be acquired by directly querying the database of the lab information system using SQL or another equivalent structured language. Results of the database query can also be placed in the same format and equivalently processed by most data analytics packages.
At this time, many laboratorians experience limitations with access to LIS data, in both timeliness and breadth. Results from ad hoc queries enable development and validation of machine learning models using retrospective data. However, data access must become timely and automated to facilitate implementation of machine learning pipelines, as discussed in a later section. Though of benefit due to its large case mix, aggregated laboratory data in multisite repositories is likely subject to method bias related to the different measurement procedures used. Such data veracity issues should be examined during exploratory data analysis and addressed prior to modeling. Finally, because not every patient gets the same panel of lab tests, some LIS datasets may be sparse (that is, many values are not available for many analytes). Sparseness is discussed in a later section of this chapter.
The data produced by mass spectrometers can come in several forms depending on the instrument and how the instrument acquisition method is designed. In this section, some of the most common types of mass spectrometry data will be described based on the type of instrument. For a more comprehensive background introduction to the principles of mass spectrometry, see Chapter 20 .
To optimally utilize mass spectrometry data, it is important to understand both the various scales and dimensions of mass spectrometry data and the methods by which mass spectrometry data can be accessed for use in machine learning applications. As a rule, the raw binary files created by instrument acquisition software systems cannot be read directly. Instrument vendors have many reasons for using a binary format, which is sometimes even encrypted, as the native format for a mass spectrometer. There are two ways to access raw data from mass spectrometers. One way is to use a software library supplied by the vendor to extract data directly from the native binary format. The second is to use one of several file formats that have been created as either formal or de facto standard file formats. Some instrument vendors provide a feature in the acquisition software to generate an export file. In this section, the most common export file formats will be discussed. Direct access to the vendor binary files will not be covered. For information on using instrument vendor software libraries, refer to the instrument vendor documentation.
Mass spectrometry data can take a 1-D (intensity of a single ion) or 2-D (intensity of multiple ions forming a spectrum) form. Instruments often report data using flat files; this is especially true for 1-D data, but flat files can be used for 2-D data as well. The most common flat file for spectra is the format called JCAMP-DX used by groups like National Institute of Standards and Technology (NIST) and the US Environmental Protection Agency (EPA). , It is relatively straightforward to read flat text files containing either 1-D or 2-D data from the types of instruments mentioned for purposes of machine learning. One of the most common applications using only 2-D spectra is the look-up of an unknown from a library. Library search requires selecting a similarity measure and is subject to the curse of dimensionality described below in the section on additional principles and limitations of machine learning. Recently, medical devices that perform microbial identification have been constructed using matrix assisted laser desorption time-of-flight (MALDI-TOF) single-stage analyzers, which use libraries of standardized spectra from a wide range of organisms to perform a probabilistic identification. ,
When some form of chromatographic separation is introduced into a mass spectrometry system, 3-D data (spectra over time) are available. The mass spectrometer is then usually configured to repeat an acquisition for a period of time. Again, the intensity of a single mass-to-charge ratio ( m/z ) value could be monitored for a period of time, which would produce a 2-D data set (sometimes called a chronogram, or a chromatogram if there is chromatographic separation) where now the x -axis is time. However, it is also typical to monitor several m/z values during the sample introduction period, which produces a three-dimensional data set representing a collection of 2-D chronograms. In this 3-D data, the x -axis is usually time, and the y -axis is used to represent the individual m/z values monitored, and the z -axis represents the intensity observed. Common applications of chronograms include inductively coupled plasma mass spectrometry (ICP-MS) analysis of metals, flow injection, more recently ambient ionization systems such as desorption electrospray ionization (DESI), and the various derivatives of paper-spray. Chromatograms are routinely used in clinical applications for a variety of analytes (see Chapter 20 ).
Some instrument software can export 3-D data in a flat-file format. While JCAMP-DX does not formally support the extra time dimension, there have been extensions proposed to allow the standard to be used for chromatography data. To represent the time dimension, a format called “analytical data interchange format for mass spectrometry” (ANDI-MS) was created by the Analytical Instruments Association (AIA) based on the Network Common Data Format (netCDF) developed and maintained by the Unidata program at the University Corporation for Atmospheric Research (UCAR). AIA/ANDI-MS was adopted as a standard by the ASTM International organization. The format has been known variously as ANDI-MS, AIA, or netCDF. The ASTM standard uses netCDF because it is a machine-independent binary file format that can hold almost any array-like data. The format can be used to hold single spectra, chronograms, and full chromatographic data. Programming libraries are needed to access netCDF, but they are not dependent on an instrument vendor. There are many language bindings for netCDF data, and the format is extremely well documented and supported by UCAR. The specific variables stored in the netCDF file according to the ASTM specification, however, are limited. The most significant limitation is the lack of a way to store higher dimensions of mass spectrometry. This is not an actual limitation of the UCAR netCDF format, but rather the way the format is used to store mass spectrometry data.
It is now common to collect clinical data using one or more liquid chromatographic stages combined with two stages of mass spectrometry. This has typically been called LC-MS/MS. For machine learning applications, the extra stage of mass spectrometry requires keeping track of the m/z value used by the first mass analyzer (precursor ion). The second stage of mass analysis can then be set, as described above, to either collect a full product ion spectrum or monitor a single product ion. When one precursor/product pair is monitored, it’s called single-reaction monitoring (SRM). When multiple m/z pairs are monitored, it is commonly called multiple-reaction monitoring (MRM). In both cases, chromatograms are generated, representing the intensity of a precursor ion fragmenting into a product ion. The ASTM netCDF format makes no provision for tracking the extra information of the product/precursor pair, so it cannot be used to store data for SRM or MRM measurements. New formats have been created, primarily based on the XML standard, to represent tandem mass spectrometry data. XML is essentially a well-defined specification for a flat text file which improves machine readability.
The first XML format to be developed for mass spectrometry data was mzXML, which is well supported and is an extremely practical format for all types of mass spectrometry data analysis. The mzXML format was developed at the Institute for Systems Biology (ISB) to support the first proteomics applications. As such, it is under the control of the ISB, which has the benefit of a very long-running support team and widespread use through the distribution of ISB open-source tools. It does, however, change as needed to support the ISB tools. It also uses some non-XML features to improve performance. To establish a more robust exchange data format, the Human Proteome Organization (HUPO) created an open, multi-vendor standards body along with processes to approve changes to the format. The standard produced is now called mzML and has the support of vendors, developers (including the ISB), and journal editors. The mzML format deliberately trades performance for completeness. Unlike mzXML, which uses a fixed collection of elements and attributes to describe complex mass spectrometry measurement, mzML uses a small set which must be combined with a controlled vocabulary. The schema for mzML has not changed in many years, ensuring that software designed to read the data will not be broken by changes. The controlled vocabulary, called psi-ms, however, is being constantly updated as new instruments and new experiments are invented. Both the format and the controlled vocabulary are under the control of an editorial board, and both are supported by a very active community. The choice to use mzXML or mzML should be determined by the application. When performance is critical, mzXML is a clear choice. When long-term interchangeability is important, mzML has a better chance of being readable over the long term.
The XML formats described above can be used for almost any mass spectrometry experiment. They are designed to handle very high-dimensional data such as the type collected for data-dependent and independent acquisition, as well as multiple stages of mass spectrometry (sometimes referred to as MS n ) and spectra collected with a variety of system settings such as multiple collision energies and other parameters.
Finally, many mass spectrometry measurements are designed to produce a quantity or ratio. For flow injection, this may be a quantity based on the intensity of a single m/z value. When collecting a single spectrum, the result could be the ratio between various m/z intensities or the presence or absence of an ion signal at a given m/z value. In chromatography measurements, the result could be everything from the area of a single SRM peak or the combination of multiple SRM peaks further processed into a quantity through the use of a calibration curve. All of these results have potential use in machine learning applications. The most common data format for mass spectrometry results is a vendor-specific flat file. These usually require complex parsing because many times, flat files are designed to be read by humans and not machines. One solution to improving the accessibility of results data is a format called mzTab-M developed by a consortium including HUPO, the Metabolomics Society, and the Metabolomics Standards Initiative. mzTab-M is a tab-delimited format that is designed to hold the results of complex mass spectrometry measurements and be easily readable by both humans and machines.
The data produced by mass spectrometers in clinical applications can range from single numbers to very high dimensional data. Through the work of many groups, and with the collaboration from instrument vendors, there are now several ways to access everything from raw spectral data to computed results. Using the tools provided by these informatics groups, it is now possible to develop sophisticated machine learning systems from almost any instrument vendor’s data at any level of complexity.
The general term “omics” refers to the high-throughput methods to comprehensively characterize and quantify a large number of molecules, grouped by their structural or functional similarities. Mass spectrometry (discussed above) is one method of generating omics-scale data; however, there are many others that also provide rich sources of data for machine learning. For example, next-generation sequencing can provide not only genomic data but also quantitative transcriptomic information through RNA-seq. In this case, every subject or sample will have a numerical result for each transcribed gene that is measured indicating its abundance.
Omics-scale data can create technological challenges because their size and complexity exceed the capacity of traditional infrastructure components used for data storage, data transfer, and computational power. An additional challenge with these datasets is that often the number of variables ( p ) measured vastly exceeds the total number of cases ( N ), and situations with p >> N can be particularly prone to overfitting (see below) and other problems. Care is required when using machine learning approaches in this setting, with a very high number of dimensions, to ensure that robust models are created.
It is a fact of statistical machine learning that there is no one best type of model for all problems. This result has come to be known as the No Free Lunch Theorem. All models have three sources of error that add up to give the total estimation error: Bias , Variance , and the Irreducible error. The first two are directly related and must be balanced to achieve a sound machine learning system. The third is independent of the first two and requires a separate approach to reduce its impact.
Bias : This is the error made by the model due to generalization. Bias is the result of the model being simpler than the real-world problem being solved or the features used by the model are not informative enough. A biased model is not totally without value. For example, in regression, we often choose a simple linear model to approximate nonlinear, or noisy data. The downside of a biased model is that, regardless of the amount of training data used, the error will not be reduced beyond some limit. Thus for highly nonlinear data, a linear model will have high bias . High bias models underfit the training data. The bias of a model can be lowered by increasing the complexity of the model or increasing the number or information content of features. An unbiased model is one for which increasing the amount of data will result in making the bias lower, ultimately to zero. However, counter-intuitively, zero bias is not always desirable. For example, in data with noise, a regression model with zero bias would draw a curve that went through every data point, which means the model has simply memorized or overfit the training data, not actually modeled it. For classification problems, this is the same as getting all of the labels on the training data precisely correct. This type of model will not perform well when new data are introduced.
Variance: This is the error caused by a model being sensitive to variations in the training data. Variance is the result of the model being more complex than the real-world problem being solved, or containing too many features for the size of the training set (see the discussion of p >> N in the previous section). A model with high variance is overly sensitive to small fluctuations in the training data and can be the source of overfitting mentioned above. It is this relationship between bias and variance that must be considered when building a model. A low-bias model with high variance will overfit the training data and perform poorly on data that is not part of the training set. A low-variance model is one that is not sensitive to fluctuations in the training data. A low variance model with high bias will underfit the training data and also perform poorly on new data.
Irreducible error: This error is due to the actual noise in the data. Noise can take many forms and should be characterized in order to design a process to lower it. There are many ways to “clean up” data before processing. Signal processing can help reduce the noise in measurement data. Care must be taken to apply methods such as signal averaging and filtering. Distinguishing signal from noise is a nontrivial task, and simplistic methods can distort data in a way that makes models perform worse than they did without noise reduction. The other primary source of irreducible error is outliers. An outlier is an observation that is an abnormal distance from other values in a random sample from a population. In statistical machine learning, many measures of model performance can be highly distorted by outliers. For example, when using least squares for either regression or classification, the mean squared error (MSE) is used, and the mean is known to be especially sensitive to outliers.
The ideal machine learning model has both low bias and low variance, and finding the right tradeoff between these two types of error is the purpose of model validation. The data available for training a model can be used in various ways to maximize the performance of a particular model. Constructing a robust validation process combined with good data cleaning is a requirement for building a model that performs well with real-world data.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here