Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The availability of rapid and reliable viral diagnostic tests, particularly nucleic acid amplification tests (NAATs), facilitates decision making in the prevention, treatment, public health, and infection control measures related to viral infections. With specific antiviral therapy available for many clinically relevant viruses, a specific viral diagnosis may limit the need for further diagnostic testing and unnecessary antibiotic therapy.
Two major approaches to diagnosis of viral infection are virologic (detection of virus) and serologic (detection of antibody, antigen, or both). The virologic approach includes (1) isolation of infectious virus in cell culture, (2) detection of viral antigen by immunologic methods (e.g., fluorescent antibody [FA] testing or enzyme immunoassay [EIA]); (3) visualization of viral particles by electron microscopy (EM); and (4) detection of viral nucleic acid by molecular techniques (e.g., hybridization or NAATs). Cytologic examination of tissues and cells may identify viral effects, thus prompting further investigation. Occasionally, cytologic changes can be sufficiently specific to suggest a particular agent (e.g., cytomegalovirus [CMV]). The serologic approach includes detection of the following: (1) virus-specific antibodies indicating recent, current, or past infection, as well as immunity following recovery or vaccination; (2) a significant rise in virus-specific immunoglobulin G (IgG) antibody suggestive of acute or recent infection; (3) virus-specific antigens (e.g., hepatitis B surface antigen [HBsAg]); or (4) virus-specific IgM antibody in late acute or early recovery phase sera. As the immune response matures following a viral infection, low-avidity IgG antibodies are replaced with high-avidity antibodies. EIAs capable of measuring the avidity of IgG antibodies to specific viruses have been used to distinguish primary from secondary (reactivation) antibody responses to vaccination or natural infection. ,
Clinically, laboratory tests for the detection of virus infection can be divided into 3 specific categories: those used to (1) make a specific diagnosis, (2) measure virus activity in patients known to be infected (e.g., viral load testing for HIV), and (3) screen for infection or a carrier state (e.g., before transplantation or blood donation).
For the detection of most viruses, specimens collected soon after the onset of clinical symptoms (preferably within the first 3–4 days) when viral shedding is greatest are preferred. Optimal specimens vary depending on the site or sites of disease. In general, tissues, aspirates, and body fluids are superior to swabs. Body sites or lesions that can be sampled easily with a swab include the pharynx or nasopharynx, conjunctiva, urethra, cervix, vagina, and vesicles or ulcers on the skin or mucous membranes. Many swab types are available for specimen collection, including plastic swabs, wooden swabs, and swabs with a flexible wire shaft and a tip made of cotton, Dacron, calcium alginate, or polyurethane, although not all are suitable for detection of some viruses. Swabs with a wooden shaft can contain toxic products that inactivate herpes simplex virus (HSV). Cotton-tipped swabs can contain fatty acids that can interfere with the survival of Chlamydia species, but they are suitable for the collection of specimens from the vagina, cervix, or urethra for the detection of Mycoplasma. Calcium alginate–tipped swabs can be toxic for lipid-enveloped viruses such as herpesviruses and some cell cultures, but they are useful for the collection of specimens for Chlamydia. Although swabs placed in a viral transport medium (VTM) can be used for NAATs, many commercial assays for detection of viruses and Chlamydia provide their own swab and transport media, which should be used.
Swabs and tissues for detection of viruses should be placed into VTM to prevent drying, maintain virus viability, and prevent the overgrowth of contaminating organisms. Several commercially prepared VTMs are available. Swabs collected for bacterial isolation that are placed in bacterial transport medium are unacceptable for detection of viruses. Conversely, VTM contains antimicrobial agents that inhibit most bacteria and fungi. Specimens such as blood, bone marrow, cerebrospinal fluid (CSF), urine, and other body fluids should be placed in clean, sterile containers without VTM.
For detection of most respiratory viruses, a nasopharyngeal (NP) aspirate or wash, sputum, or bronchoalveolar lavage (BAL) specimen provides a better yield than NP, nasal, or throat swabs. Multiple samples can be required to maximize yield. Freshly passed stool is superior to a rectal swab for detection of gastrointestinal viruses.
Specific viruses can be found in different blood cells, the plasma or serum, or both (e.g., HIV in lymphocytes and macrophages, CMV in neutrophils and to a lesser extent in mononuclear cells, enteroviruses in plasma and white blood cells). , Blood should be collected into Vacutainer tubes containing an anticoagulant such as ethylenediaminetetraacetic acid (EDTA). Recovery rates are higher with EDTA than with heparin. Heparin can inactivate herpesviruses and can inhibit some NAATs , ; this issue may be less of a concern for real-time polymerase chain reaction (PCR) and can be related to the type of heparin (sodium vs. lithium) used. ,
For tissue specimens or when the lability of particular viruses (e.g., respiratory syncytial virus [RSV] or varicella-zoster virus [VZV]) is a concern, VTM containing albumin or serum as a stabilizer should be used.
Most viruses are stable for 2–3 days at 4°C (refrigerator or wet ice temperature). Freezing at −20°C (ordinary freezer temperature) destroys or reduces the infectivity of most viruses and can alter the ability to detect viral antigen when using some commercially available kits. Beyond 2–3 days, specimens should be stored in an ultralow-temperature freezer (−70°C) and transported on dry ice. For some NAATs (e.g., detection of hepatitis C virus [HCV] RNA in serum or plasma), serum or plasma should be separated within 4–6 hours of collection and processed within 72 hours (if kept at 2°C–8°C) or frozen at −70°C until tested.
For serologic detection of viral antibodies or antigen, blood can be transported at room temperature. If a delay is anticipated, the specimen should be kept refrigerated at 2°C–8°C. Serum or plasma should be separated as soon as possible after specimen collection. If an extended period will elapse before testing, the serum or plasma sample should be frozen at −20°C or lower. Repeated freeze-thaw cycles should be avoided. For viruses for which an IgM assay is available (e.g., hepatitis A virus [HAV]), an acute phase specimen can be sufficient for diagnosis. Otherwise, an acute phase specimen is collected within a few days of illness onset and serum is stored, followed by collection of a convalescent phase specimen 2–4 weeks later, and the specimens are tested simultaneously.
Most laboratories performing virus isolation use monolayer cell culture techniques. However, many clinically relevant viruses, such as parvovirus, human papillomavirus, hepatitis viruses, Epstein-Barr virus (EBV), rotaviruses, noroviruses, and others, are not cultivatable in routine diagnostic laboratories; laboratory diagnosis is based on other methods. Although cultivation of HIV is possible using suspension cultures of lymphocytes, special containment facilities are required; alternative methods are used for routine diagnosis. The major viruses detected by isolation in monolayer cell culture include HSV-1 and HSV-2, CMV, VZV, RSV, influenza A and B viruses, parainfluenza viruses, respiratory adenoviruses, several enteroviruses (coxsackievirus, echovirus, poliovirus), and measles virus. Not all cultivatable viruses replicate in a single cell line. Thus various cell lines are used for primary isolation: diploid cell lines such as human foreskin or lung fibroblasts for herpesviruses, primary cell lines such as primary rhesus monkey kidney cells for respiratory viruses and enteroviruses, and heteroploid or continuous human epithelial cell lines such as Hep-2 cells for RSV. The types of cell lines used in the diagnostic laboratory are determined by the specimen type, season, epidemiologic data, and clinical information provided. Many viruses cause morphologic changes (i.e., cytopathic effect[(CPE]), in the cell monolayer. Some viruses cause CPE within 2 days (e.g., HSV), others within a week (e.g., enteroviruses), and others after several weeks (e.g., CMV). For viruses that do not cause a typical CPE, detection can be based on the adsorption of red blood cells to the surface of virus-infected cells in culture (e.g., influenza and parainfluenza viruses) or by the use of interference assays (e.g., rubella virus). Presumptive identification of a particular virus or virus group in cell culture is based on the cell type, the timing and appearance of CPE, the source of the specimen, and the suspected clinical diagnosis.
Confirmation of the virus isolated requires immunologic methods such as fluorescein- or peroxidase-conjugated virus-specific monoclonal and polyclonal antibodies. Antibodies to HSV, CMV, VZV, RSV, influenza A and B virus, parainfluenza virus, adenovirus, measles virus, and enterovirus antigens are readily available.
Centrifugation of specimens (referred to as shell vial or spin-amplified culture) onto cell monolayers on coverslips placed in the bottom of small vials or in wells, followed by incubation and staining for viral antigen by using monoclonal antibody after 1–3 days, shortens the time required to detect and confirm the presence of many viruses and has replaced conventional cultures in many laboratories. For slowly growing viruses such as CMV, the use of monoclonal antibody against nonstructural proteins produced early in the replication cycle (i.e., immediate early antigen [EA] or EA) allows detection of virus days to weeks before CPE can be observed by traditional cell culture techniques.
Two techniques for isolation of some viruses have been developed with sensitivity comparable to that of standard culture and shell vial methods. Genetically engineered cell lines such as the enzyme-linked virus-inducible system (ELVIS, Quidel, San Diego, CA) using a baby hamster kidney cell line that has been transformed by using an HSV-inducible promoter ( UL39 gene) and an Escherichia coli β-galactosidase gene was introduced for the isolation of HSV. The addition of a substrate for the β-galactosidase enzyme results in formation of a color reaction in the HSV-infected cells. This technique has been adapted for performing rapid HSV antiviral susceptibility testing. Mixing multiple cell types in a single shell vial culture can provide rapid detection of the following: respiratory viruses (R-Mix, Quidel); HSV, CMV, and VZV (H&V-Mix, Quidel); and enteroviruses (E-Mix, Quidel).
Virus antigen detection tests can be performed directly on a variety of specimen types and are highly specific and rapid. Because virus antigen is cell associated, collection of an adequate number of infected cells is important. Several commercial kits (EIA, latex agglutination, FA) are available for the detection of the following: (1) rotavirus and enteric adenovirus in stool specimens; (2) RSV, influenza A and B viruses, parainfluenza viruses, and adenoviruses in respiratory tract specimens; (3) HBsAg and HIV p24 antigen in serum; (4) HSV and VZV in vesicle or ulcer swab specimens; and (5) CMV in BAL and blood specimens. The FA technique has been used for the detection of rabies virus in brain tissue, mumps virus in throat and urine sediment, and measles virus in conjunctival cells. The detection of CMV pp65 antigen in neutrophils can be used in the diagnosis and management of new or reactivated CMV infection in immunocompromised patients.
Although almost any specimen type is suitable for EM, most laboratories have replaced EM with other virus detection methods. , Some laboratories continue to use EM for the detection of gastrointestinal pathogens such as rotavirus, enteric adenoviruses, and norovirus, as well as for detection of BK-polyomavirus in tissue biopsy and urine samples. Disadvantages of EM include the large number of virus particles (approximately 1 × 10 6 /mL of specimen) required for detection, limited throughput, expense, and lack of availability and expertise in many centers.
The increased sensitivity and availability of NAATs for detection of almost any clinically relevant virus have revolutionized testing in the clinical virology laboratory. , In many laboratories, NAATs have supplanted virus culture, antigen detection methods, and direct molecular hybridization techniques. Three approaches have been taken: (1) target amplification such as PCR, strand displacement amplification, nucleic acid sequence–based amplification, and transcription-mediated amplification systems; (2) probe amplification, including Q-β replicase and ligase chain reaction; and (3) signal amplification, such as branched-chain DNA (bDNA) assay and hybrid capture assay. , Several commercial and in-house (“home-brew”) assays have been developed. Quantification of viral genome in plasma or serum can be used to determine prognosis, select patients for antiviral therapy, and monitor response to treatment in a variety of patient populations. , Multiplex assays capable of detecting several viruses in a single amplification reaction have been developed (e.g., for herpesviruses and for enteric, bloodborne, and respiratory viruses), and studies suggest that these assays are cost-effective. The development of automated real-time PCR using fluorescence techniques and continuous detection of amplified product has shortened detection times substantially relative to conventional PCR assays. Because these assays use a closed system (i.e., amplification and detection occur in a single tube that need not be opened once the reaction is completed), they also are less prone to contamination than conventional PCR. NAAT has been applied to genotyping of viruses (e.g., HIV, HBV, and HCV), as well as for the detection of mutations that confer resistance to antiviral agents. Newer molecular techniques such as pyrosequencing and whole-genome sequencing are beginning to be applied in the clinical laboratory and are providing improved genotyping of viruses, detection of virus quasispecies, and metagenomic analysis for detecting existing and new or novel viruses in clinical specimens. ,
Choosing optimal tests depends on the specific virus being sought, clinical setting, specimen type, availability of kits, reagents and equipment, experience of laboratory personnel, and cost. The use of NAATs is rapidly replacing older viral diagnostic methods because of the rapid turnaround time, superior sensitivity, and ability to quantify virus density of NAATs. Several relatively simple in-house and commercially available NAATs are available for a wide variety of viruses. However, some laboratories continue to use antigen detection methods and virus culture when expertise in molecular techniques is not available or cost is prohibitive.
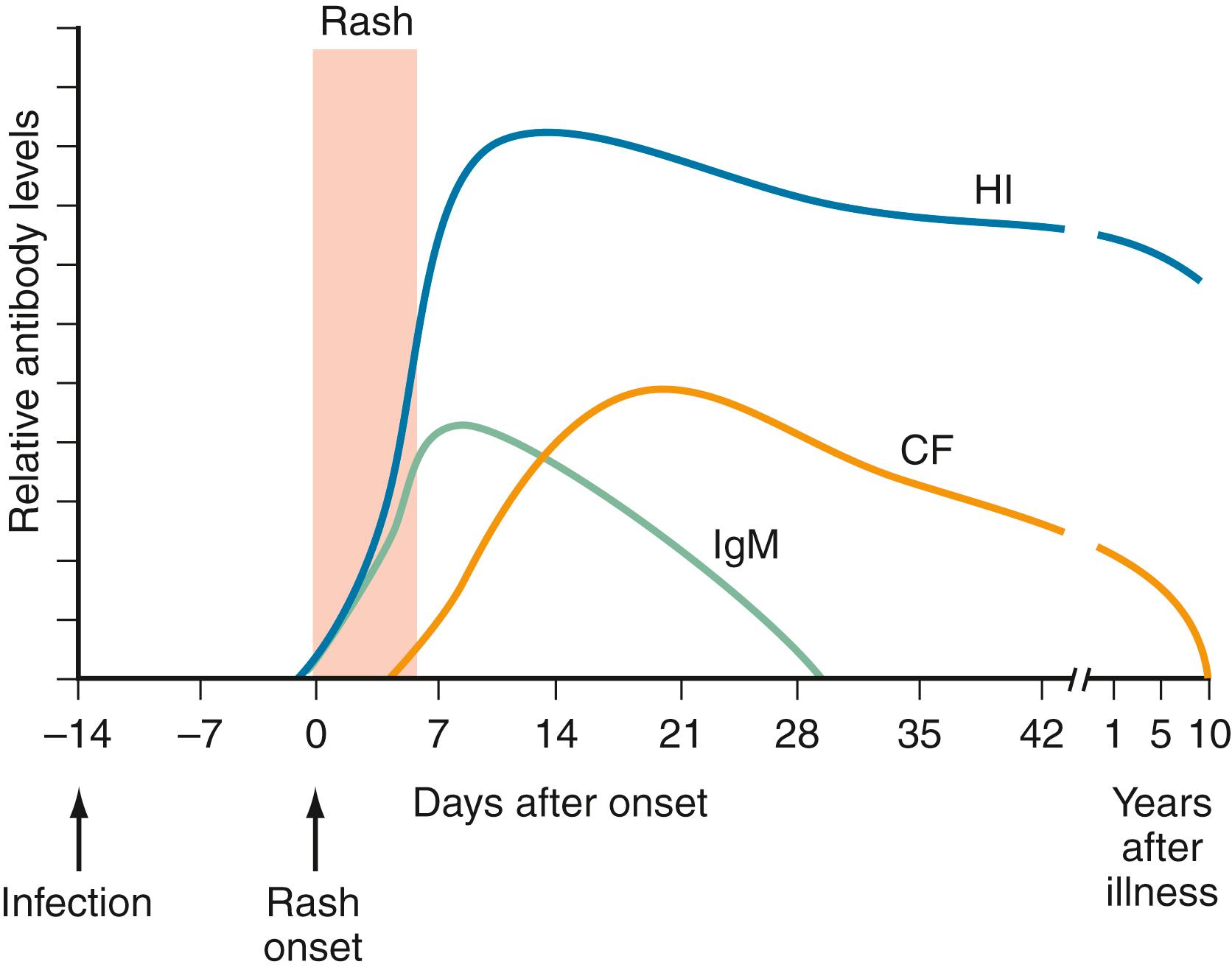
Serologic methods can be used to diagnose a current or recent acute infection, to determine specific susceptibility or immunity, and for epidemiologic and surveillance purposes. Interpretation of serologic results is virus specific. For example, the presence of HIV antibodies indicates current infection, whereas the presence of IgG antirubella indicates immunity as a result of immunization or recovery from natural infection. Serologic diagnosis of acute infection is more useful when the incubation period is prolonged (e.g., 3–6 weeks) and antibody is present in serum concomitantly with signs of illness (e.g., EBV and CMV mononucleosis). Fig. 287.1 shows a typical antibody response for an acute, moderate-incubation (several days to 2 weeks) viral illness such as measles. At the onset of rash or other manifestations, antibody is undetectable or is present at low titer. Within 10–14 days, appreciable titers of antibody are present. For short-incubation virus infections (e.g., respiratory viruses), a rise in antibody usually does not occur until the late recovery phase or during convalescence and therefore has no value for diagnosis during acute infection. Older serologic methods such as hemagglutination inhibition and complement fixation (CF), which relied on a greater than fourfold rise in antibody titer between acute and convalescent sera tested in parallel, have largely been replaced by solid phase immunoassays such as EIAs, passive latex agglutination, and immunofluorescence assay (IFA). The presence of antibody in high titer in a single serum specimen during convalescence usually does not permit a definitive diagnosis. Seroconversion can be used to diagnose an acute or recent infection.
EIAs and enzyme-linked immunoassays (ELISAs) are highly standardized, can be quantitative, can detect IgG or IgM antibodies or both, and, for some viruses such as HIV and HCV, can simultaneously detect both antigen and antibodies in a single assay. , EIAs can be noncompetitive (“sandwich assay”) or competitive assays. Results usually are measured in optical density (OD) units because of a color reaction, and results are reported either qualitatively or quantitatively in international units (IU) or index values. Interpretation of optical density units varies with the EIA-ELISA kit used and the virus antibodies being detected. A relatively newer modification of EIA (known as chemiluminescent immunoassay) uses an enzyme/substrate combination to generate a light signal and results are reported as relative light units (RLU). Chemiluminescent immunoassays generally are considered more sensitive than EIAs.
The presence of virus-specific IgM antibody in serum obtained 1–2 weeks after the onset of illness permits a diagnosis of acute or recent infection for many viruses. Typically, IgM antibody disappears from serum within a few months after the acute illness, but it can persist for an extended time in some people and for some viruses. False-positive IgM results can be caused by (1) cross-reactivity (e.g., among herpesviruses or from polyclonal stimulation secondary to EBV infection), (2) the presence of rheumatoid factor (IgM antibody that binds to the Fc portion of IgG), and (3) inherent testing difficulties. Misinterpretation of IgM antibodies as indicative of an acute or recent infection also can occur as a result of (1) persistence of IgM antibody for several months after the acute illness (e.g., EBV, West Nile virus) or (2) reactivation of latent or chronic viruses (e.g., HSV, HBV).
False-negative IgM test results can reflect the following: (1) an absent, low, or delayed IgM response, especially in immunologically immature hosts (e.g., during infancy, congenital CMV, or HIV infection) or in immunosuppressed patients (e.g., patients with AIDS) , or (2) the presence of high-titer IgG antibody (precluding binding of IgM). Many commercially available kits contain reagents to adsorb IgG from the test serum or use a background subtraction step, thus reducing the possibility of interference. ,
When IgG antibody tests are used to determine susceptibility or immunity to a particular virus, the sensitivity of the method is important. Detection of neutralizing antibodies can be the best predictor of immunity.
The major advantages of serologic diagnosis of acute viral infection include noncritical specimen handling and wide availability. The disadvantages include (1) the requirement for acute and convalescent sera for IgG antibody tests, (2) false-positive and false-negative IgM antibody results, and (3) a delay of 2–3 weeks before a diagnosis can be confirmed with short-incubation infections. Because of the many confounding factors (e.g., passive transfer of antibodies from mother to infant, receipt of immunoglobulin, immunocompromise), serologic test results always should be interpreted within the context of the clinical situation. Whenever possible, serologic diagnosis should be confirmed with the use of virus isolation or direct detection of virus antigens or nucleic acids.
Depending on the serologic assay, either serum or plasma can be used. The use of other specimen types has not been well validated for most viruses or assays. Some exceptions include the use of saliva for the detection of HIV antibodies and CSF in patients with viral central nervous system (CNS) disease. ,
Table 287.1 contains a list of the medically important viruses, major attributable diseases, optimal diagnostic specimens, available tests, and average time to a positive test result. For many tests, the time to obtain a result is a function of the test itself (e.g., culture), the logistics of laboratory testing schedules, or the need to refer a sample to a reference laboratory. The preferred test provides the fastest result with acceptable sensitivity (>90%) and specificity (>95%). The preferred diagnostic test or tests can vary, depending on the patient population being tested (e.g., immunocompromised hosts) and the clinical indication.
| Agent/Type or Site of Infection or Host | Major Diseases | Optimal Specimens | Available Tests a | Average Test Time to Positive Results b |
|---|---|---|---|---|
| ADENOVIRUS | ||||
| Respiratory | Pharyngitis, pneumonia, undifferentiated febrile illness | NP aspirate or wash, NP swab, throat swab, BAL, lung tissue | Culture c | 6 days |
| PCR | 1–2 days | |||
| Antigen detection/FA | 2 hr | |||
| Serum | IgG antibody d | 1–5 days | ||
| Eye | Conjunctivitis | Conjunctival swab or scraping | Culture c | 7 days |
| Antigen detection | 2 hr | |||
| Serum | IgG antibody c | 1–5 days | ||
| Intestinal (types 40 and 41) | Gastroenteritis | Stool | Antigen detection | 2 hr |
| EM | 2 hr | |||
| Urinary bladder (immunocompromised host) | Hemorrhagic cystitis | Urine | Culture | 6 days |
| PCR c | 1–2 days | |||
| EM | 2 hr | |||
| ARBOVIRUSES | ||||
| SLE, California, WEE, EEE, WNV | Fever, meningoencephalitis | Serum, CSF | IgG and IgM antibody d | 1–5 days |
| Colorado tick fever | Fever, malaise, neutropenia | Serum | IgG antibody | 7 days |
| Dengue | Febrile illness with or without rash, hemorrhagic fever | Serum | IgG and IgM antibody c | 1–5 days |
| PCR | 1–2 days | |||
| CHLAMYDIA | ||||
| Chlamydia trachomatis | ||||
| Genital | Urethritis, proctitis, cervicitis, salpingitis, pelvic inflammatory disease | Urethral, cervical swab, first-void urine, self-collected vulvovaginal swab, rectal mucosal swab | NAAT c | 2–6 hr |
| Antigen detection | 4 hr | |||
| DNA probe | 4 hr | |||
| Culture | 48–72 hr | |||
| Neonatal | Conjunctivitis, pneumonitis | Eye swab, NP aspirate or wash | NAAT c | 2–6 hr |
| Antigen detection | 4 hr | |||
| Culture | 48–72 hr | |||
| Sexual abuse, rape | Vaginitis, urethritis, proctitis | Cervical, urethral, rectal mucosal swab | Culture e | 48–72 hr |
| Chlamydia (Chlamydophila) pneumoniae | Pneumonia, pharyngitis, bronchitis | NP aspirate or swab, throat swab or wash | Culture c | 4 days |
| Antigen detection | 4 hr | |||
| Serum | IgG and IgM antibody | 1–5 days | ||
| Chlamydia psittaci | Pneumonia | NP aspirate or wash, throat swab or wash | Antigen detection | 4 hr |
| Culture | 2 days | |||
| Serum | IgG antibody d | 1–5 days | ||
| CYTOMEGALOVIRUS | ||||
| Congenital | Hepatosplenomegaly, thrombocytopenia, microcephaly, hearing loss, chorioretinitis | Urine, throat swab, EDTA blood, serum, amniotic fluid | Shell vial culture with antigen stain c | 2 days |
| Culture | 3–4 wk | |||
| NAAT f | 2–5 hr | |||
| IgG and IgM antibody c | 1–2 days | |||
| Postnatal infection | Heterophile-negative infectious mononucleosis | Throat swab, urine, EDTA blood | Shell vial culture with antigen stain c | 2 days |
| Culture | 3–4 wk | |||
| Serum | IgG and IgM antibody d | 1–2 days | ||
| Immunosuppressed patients | Pneumonitis, colitis, retinitis | EDTA blood | Antigenemia assay c | 4–6 hr |
| NAAT c , f | 2–5 hr | |||
| Bronchoalveolar lavage, rectal swab, vitreous fluid, tissue biopsy | Shell vial culture with antigen stain c | 2 days | ||
| Culture | 3–4 wk | |||
| NAAT c , f | 2–5 hr | |||
| Pretransplant screening or immune status | Past infection (donor and recipient) | Serum | IgG antibody | 1–2 days |
| ENTEROVIRUSES | ||||
| Coxsackieviruses groups A and B, echovirus, poliovirus | Aseptic meningitis, fever and rash, herpangina, hand, foot, and mouth disease, myocarditis and pericarditis, paralytic disease | CSF, throat swab, stool, rectal swab, EDTA blood, pericardial fluid, myocardium | Culture | 4–7 days |
| PCR c , f | 6 hr | |||
| Serum | Neutralizing d , g antibody panel (coxsackievirus group B, echovirus, and poliovirus) | 5 days | ||
| EPSTEIN-BARR VIRUS | ||||
| Healthy person | Mononucleosis syndrome | Serum | Slide agglutination test (monospot) c | 1–3 days |
| EBV-specific IgG and IgM antibody d | 1–3 days | |||
| Immunocompromised | Posttransplant lymphoproliferative disease | Serum, plasma, whole blood, leukocytes | PCR (quantitative) f | 2–5 hr |
| GASTROINTESTINAL VIRUSES | ||||
| Rotaviruses, Caliciviridae (norovirus and sapovirus), enteric adenoviruses, astroviruses | Gastroenteritis | Stool | EM c (rotavirus and enteric adenovirus) c | 2 hr |
| PCR f | 6 hr | |||
| GENITAL MYCOPLASMA AND UREAPLASMA | ||||
| Ureaplasma urealyticum | Urethritis, cervicitis | Urethral, cervical swab; semen | Culture c | 2 days |
| Mycoplasma hominis | Pneumonitis, meningitis in neonates | Tracheal aspirate, CSF in neonates | Culture c | 2 days |
| Mycoplasma genitalium | Urethritis, cervicitis | Urethral, cervical swab | Culture NAAT | ≥6 days 6 hr |
| HEPATITIS VIRUSES | ||||
| Hepatitis A | Acute | Serum | IgM antibody | 1–2 days |
| Immunity | Serum | Total (IgG and IgM) antibody or IgG antibody | 1–2 days | |
| Hepatitis B | Acute | Serum | HBsAg, anti-HBc IgM | 1–2 days |
| Chronic | Serum | HBsAg, anti-HBc total antibody | 1–2 days | |
| Serum or plasma | NAAT for HBV DNA (quantitative) f | 1 week | ||
| Immunity | Serum | HBsAb | 1–2 days | |
| Hepatitis C | Acute | Serum | Anti-HCV EIA screen | 1–2 days |
| Anti-HCV RIBA supplementary | 5 days | |||
| Chronic | Serum or plasma | NAAT for HCV RNA (quantitative or qualitative) f | 1 wk | |
| Hepatitis D (only occurs in patients with HBV coinfection/superinfection) | Acute | Serum | HDV Ag, anti-HDV IgM | 1–8 days |
| Chronic | Serum | HDV Ag, anti-HDV total | 1–8 days | |
| Hepatitis E | Acute | Serum | IgG and IgM antibody | 1–8 days |
| HERPES SIMPLEX VIRUS | ||||
| Skin, mucous membranes | Oral, genital, cutaneous ulcers or vesicles, herpetic whitlow | Aspirate of vesicle fluid, swab of vesicle fluid or base of ulcer in VTM | Shell vial culture with antigen stain c , h | 16–24 hr |
| Antigen detection (FA) | 2 hr | |||
| NAAT f | 2–5 hr | |||
| Past infection | Recurrent genital symptoms but culture results negative | Serum | IgG (group- or type-specific) antibody d | 1–2 days |
| Neonatal infection | Disseminated disease; hepatitis; pneumonitis; encephalitis; skin, eye, mouth ulcers or vesicles | Swab of lesions; EDTA blood; CSF; conjunctiva, nose, or mouth swab; rectal swab | Shell vial culture with antigen stain c , h | 16–24 hr |
| Antigen detection (FA) | 2 hr | |||
| PCR | 2–5 hr | |||
| Serum | IgG and IgM antibody d | 1–2 days | ||
| Ocular infection | Conjunctivitis, keratitis | Conjunctival or corneal swab or scraping in VTM | Shell vial culture with antigen stain c , h | 16–24 hr |
| Antigen detection (FA) | 2 hr | |||
| PCR | 2–5 hr | |||
| Brain or meninges | Encephalitis, i meningitis | CSF, brain biopsy i | PCR c , f | 2–5 hr |
| Antigen/antibody in CSF | 2 hr | |||
| Shell vial culture with antigen stain h | 16–24 hr | |||
| Serum | IgG and IgM antibody d | 1–2 days | ||
| HUMAN HERPESVIRUS 6 | ||||
| Primary infection | Roseola (exanthem subitum) | Serum | IgG and IgM antibody d | 1–3 days |
| Immunocompromised people | Transplant recipients, AIDS | EDTA blood for PBMCs | PCR f | 1–2 wk |
| HIV | ||||
| Suspected HIV infection in adult or older child | Symptomatic or asymptomatic | Serum | Screening HIV EIA c | 1–2 days |
| Confirmatory Western blot or IFA | 1–3 days | |||
| HIV p24 antigen, NAAT i | 2–4 days | |||
| Newborn | Suspected vertical or perinatal transmission | Serum | Screening HIV EIA | 1–2 days |
| Confirmatory Western blot or IFA | 1–3 days | |||
| EDTA blood | Virus culture | 2–3 wk | ||
| NAAT c , f , i | 1 wk | |||
| OTHER VIRUSES | ||||
| Human metapneumovirus | Upper respiratory illness, bronchiolitis, pneumonia, croup | NP aspirate or wash, nasal or throat swab, BAL | PCR (including multiplex assays for respiratory viruses) f | 1 day |
| Human papillomaviruses | Cervical dysplasia | Cervical swab | RNA probe, hybrid capture, PCR | 1–4 days |
| Influenza viruses | “Flu” syndrome, pneumonia | NP aspirate, wash, or swab; throat swab or wash; BAL | PCR (including multiplex assays for respiratory viruses) c , f | 1 hr–1 day |
| Antigen detection for influenza A and B | 30 min–2 hr | |||
| Culture b | 7–9 days | |||
| Measles virus | Measles | NP aspirate or wash | Culture c | 5 days |
| Throat swab | Antigen detection c | 2 hr | ||
| Serum | IgG and IgM antibody d | 1–2 days | ||
| Mumps virus | Parotitis, aseptic meningitis, meningoencephalitis | Urine, throat swab, saliva, CSF, blood | Culture | 8 days |
| Serum | IgG and IgM antibody d | 1–2 days | ||
| Parainfluenza viruses | Croup, pneumonitis, bronchiolitis | NP aspirate or wash | Culture c | 4–7 days |
| Antigen detection using FA | 2 hr | |||
| Parvovirus B19 | Erythema infectiosum | Blood, serum, bone marrow, amniotic fluid cells, placental tissue, cord | IgG and IgM antibody d | 2 days |
| Aplastic crisis, congenital, hydrops fetalis | PCR | 2 days | ||
| Polyomavirus (JC and BK) | JC virus: progressive multifocal leukoencephalopathy (PML) | CSF | PCR | 1 wk |
| BK virus: polyomavirus-associated nephropathy | Urine | PCR (quantitative) | 1 wk | |
| Rabies virus | Encephalomyelitis Immune status after vaccination |
Postmortem CNS tissue, antemortem nuchal biopsy | Direct antigen detection (DFA, IHC, DRIT) c | 24–72 hr |
| Serum, CSF | IgG and IgM antibody d (IFA) | 2 wk 2 wk |
||
| Saliva (antemortem) | Culture | 24–72 hr | ||
| Serum, CSF | RT-PCR | 2 wk | ||
| Respiratory syncytial virus | Bronchiolitis, pneumonia, croup | NP aspirate, wash, or swab; throat swab or wash; BAL | Antigen detection c | 15 min–4 hours |
| Shell vial with antigen staining | 16–48 hr | |||
| Culture | 3–7 days | |||
| PCR (including multiplex assays for respiratory viruses) | 1 hr–1 day | |||
| Rhinovirus | Common cold | NP aspirate or wash | Culture | 7 days |
| Rubella | Acquired or congenital rubella | Serum | IgG and IgM antibody d | 1–2 days |
| Throat swab | Culture | 5–7 days | ||
| SARS-CoV-2 | COVID-19 | Nasopharyngeal swab; BAL or tracheal aspirate fluid | RT-PCR, other NAATs, antigen detection | 2 hr–1 day |
| VARICELLA-ZOSTER VIRUS | ||||
| Skin, disseminated | Chickenpox, herpes zoster, occasional CNS complications | Vesicle fluid, scraping of base of vesicle in VTM, CSF | Antigen detection c | 2 hr |
| Culture | 3–7 days | |||
| PCR c , f | 1 day | |||
| Serum | IgG and IgM antibody d | 1–2 days | ||
| Immune status | Past infection or vaccination | Serum | IgG antibody | 1–2 days |
| Mycoplasma pneumoniae | ||||
| Pneumonia, pharyngitis, Stevens-Johnson syndrome, meningoencephalitis | Throat swab | Culture | 3 wk | |
| CSF | PCR f | 4–6 days | ||
| Serum | IgG and IgM antibody c | 1–5 days | ||
a Available tests may vary by laboratory. Samples may need to be sent to a reference laboratory for some tests. Not all tests need to be performed in all patients.
b The average time to a positive result may be as much a function of the test itself (e.g., culture) as it is the frequency with which the test is performed in the laboratory.
c Preferred test on the basis of sensitivity, specificity, and short time to a positive test result.
d Acute and convalescent (2–4 weeks after the onset of illness) serologic testing is recommended for most viruses. IgM antibody testing is available for CMV, EBV, HAV, HBVcore, HSV, measles, mumps, parvovirus B19, rubella, and varicella-zoster virus.
e In cases of sexual abuse or rape, culture is recommended because of concern about false-positive results with nonculture methods.
f PCR test times to a positive result vary.
g In the echovirus neutralizing antibody panel, 4–5 of the most prevalent recent serotypes are chosen for the panel.
h Serotyping of the isolate as HSV-1 or HSV-2 is available.
i Detection of proviral DNA after PCR amplification may be the preferred test in young infants, in adults with mononucleosis syndrome before seroconversion, and in adults with an indeterminate Western blot result.
The optimal tests for diagnosis of the members of the Herpesviridae family vary by virus type. For suspected mucocutaneous lesions caused by HSV or VZV, clinical diagnosis is usually sufficient. If laboratory confirmation is required, an aspirate or swab of the vesicular fluid or scraping of the ulcer base placed in VTM is recommended. Other potentially useful samples include blood in EDTA when viremia is suspected (e.g., neonates), CSF in a sterile container when meningitis or encephalitis is suspected, conjunctival swab or corneal scrapings in VTM in suspected cases of herpes keratitis, and tissue biopsy in VTM or frozen (e.g., disseminated HSV and VZV in neonates or immunocompromised patients). In infants, duodenal aspirates also can be collected for suspected neonatal HSV infection. Tests for isolation of virus, direct detection of viral antigen in cells using FA staining (and monoclonal antibodies specific for HSV-1, HSV-2, and VZV), and detection of HSV and VZV DNA by NAATs are all available.
The yield on culture is quite variable and depends on the culture cell type used, the stage of the clinical infection (yield is greatest ≤4 days of onset), and the specimen type and transportation time (preferably ≤12 hours of collection). Visualization of CPE in a sensitive cell line detects 50% of HSV-positive specimens in 24 hours, 80% in 48 hours, and 95% in 72 hours. The shell vial method permits detection of HSV with 66%–99% sensitivity and 100% specificity by 16–24 hours. , ELVIS has a sensitivity similar to that of both standard and shell vial culture.
Direct detection of VZV antigens by FA of smears from lesions is more sensitive than culture. Vesicular fluid, although good for culture, is inadequate for FA testing because of the lack of cellular material.
Direct antigen detection tests for HSV have variable sensitivity (47%–95%) and specificity (85%–100%). , None is sufficiently sensitive to detect asymptomatic shedding reliably.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here