Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Cytogenetics is the study of chromosomes and the clinical application of chromosome analysis for diagnostic purposes. This chapter will deal with traditional cytogenetics, which involves examination of metaphase chromosomes.
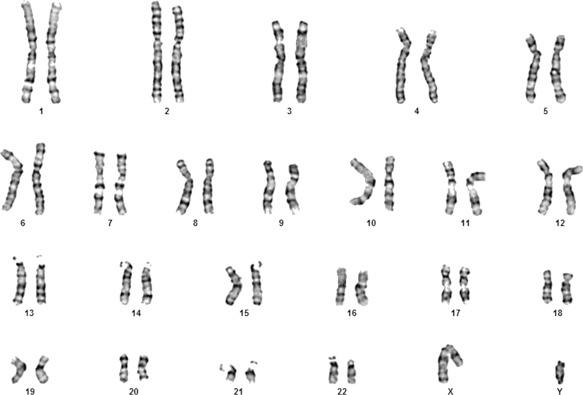
The normal euploid or diploid chromosome constitution in human cells is 46. These are present in the nucleus of the cell as 23 pairs. Germ cells in the gonads with the normal diploid chromosome constitution produce haploid gametes with 23 chromosomes; one member from each pair is contributed to the gamete, as a result of meiotic division. The diploid number of chromosomes is reconstituted at fertilization, and as a result of this process, the members of each chromosome pair are biparentally inherited. Twenty two of the pairs, known as autosomes, are numbered based on size and position of a primary constriction termed a centromere. The location of the centromere is used to classify chromosomes as metacentric (centromere at or near the middle of the chromosome), submetacentric (centromere dividing the chromosome into a short arm and long arm that are unequal in size), or acrocentric (centromere at or near one end of the chromosome). The 23rd pair is the sex chromosomes that are usually XX in females or XY in males ( Fig. 4.1 ). Chromosomes containing the nuclear DNA may be subject to partial or complete copy number variations, resulting in aneuploidy with phenotypic changes of clinical importance. In contrast to nuclear chromosomes, maternally inherited DNA in the mitochondria is not visualized by cytogenetic testing but is studied by molecular methods. Similar to nuclear DNA, mitochondrial DNA is subject to copy number variation and mutations that may be associated with phenotypic effects and diseases (see Chapter 15, Chapter 2 ).
Chromosome testing is performed to ascertain whether there is the normal number of chromosomes in the somatic cells of an individual or fetus and to detect abnormalities of chromosome structure that may have clinical or reproductive consequences.
In the cytogenetic laboratory, chromosomes are examined microscopically after staining and fixing cells on a glass slide. A standard cytogenetic test usually includes examination of clusters of chromosomes in 20 cells undergoing metaphase division. Chromosomes are physically spread on a slide by several different methods; ideally, spreading is sufficient to minimize overlapping of chromosomes but controlled to the extent that intact metaphases can be identified by electronic scanning and evaluated by experienced laboratory staff. This minimizes preparation artifact because of loss of chromosomes from one metaphase or gain of chromosomes from nearby metaphases.
Methods to differentially stain areas of chromosomes are referred to as banding. These staining methods enable identification of individual chromosomes by creating internal patterns in the members of the chromosome pairs that are common in the general population. Pretreatment with trypsin before staining with Giemsa solution results in typical staining patterns known as G-bands. G-banding is the most commonly performed banding method and produces a recognizable pattern of alternating dark and clear (light) bands, which distinguishes each chromosome pair. Some variation in size and/or staining of certain chromosome regions is observed with sufficient frequency in the general population to be categorized as normal variants or heteromorphisms. A number of staining methods other than G-banding result in patterns that may provide information about specific chromosome regions. However, these are rarely used since the advent of newer molecular methods such as microarray.
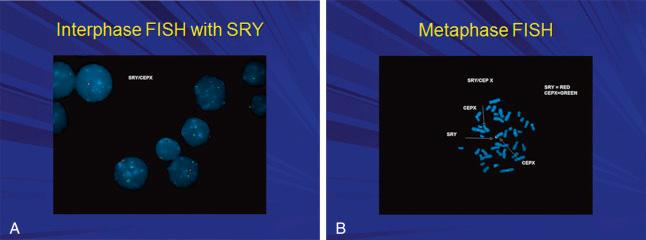
FISH uses fluorescent DNA probes that bind to regions of a chromosome that have a high degree of sequence complementarity with the probe. Upon binding, the FISH probe for a particular chromosome region will emit specific color fluorescence. Fluorescence microscopy is used to identify binding of the probe to the nucleus of interphase cells or to the metaphase chromosomes in cells undergoing mitosis. FISH on interphase nuclei may be used for rapid detection of copy number of chromosomes in cells from amniotic or chorionic villus samples when a specific trisomy is suspected. FISH probes can also be used to detect duplications, deletions, or structural rearrangement of chromosome regions, such as balanced or unbalanced reciprocal translocations, based on the locations of signals in the metaphase chromosomes (see Fig. 4.2 ).
Clinical chromosome analysis is performed on cells that are at the metaphase stage of mitotic division when there is compaction of the chromosomes into microscopically visible structures. The testing is most commonly performed on peripheral blood samples. Following addition of a mitogen to induce division of lymphocytes, exposure to an agent that arrests metaphase (e.g., colcemid) enables analysis of chromosomes in peripheral lymphocytes. Chromosomes may also be examined from cells undergoing tissue culture that have been established from biopsies of skin or other organs, including placental fragments from chorionic villus samples and desquamated cells in amniotic fluid sampling.
Tissue biopsies obtained at the time of early pregnancy loss, after stillbirth or postmortem, can often be successfully grown in tissue culture. The outer limit of time for possible growth of biopsies in tissue culture following pregnancy loss or fetal death in utero has not been defined. Chromosomal microarray (CMA) on DNA extracted from fetal tissue is recommended as the first-line test in cases of fetal death, in part because avoiding tissue culture eliminates culture failures and maternal cell contamination (see Chapter 12 ). When traditional chromosome analysis is performed tissue culture success is maximized by obtaining samples under sterile conditions and placing them in sterile containers with tissue culture media or balanced salt solution to prevent drying. When in doubt regarding the viability of the cells, it is appropriate to submit samples rather than forego indicated testing.
Short-term growth of amniotic cells or fragments of placental villi is preferred for prenatal diagnosis of chromosome abnormalities to obtain results as rapidly as possible and to minimize abnormalities or artifacts that arise in cell division during tissue culture. Laboratory standards are available regarding the processing and methods for tissue culture of samples. Counting and examining individual chromosomes in a total of 20 cells from two different tissue culture flasks or examination of chromosomes in 15 patches of cells grown on coverslips in petri dishes is recommended for prenatal diagnosis. The short-term nature of the tissue culture process on coverslips also means that there are fewer cell divisions before harvesting and consequently, less opportunity for introduction of in vitro chromosome errors.
Some tissues, such as first-trimester placenta, are growing so rapidly that there are sufficient spontaneous mitoses that can be arrested in metaphase division without initiating tissue culture. Chromosome analysis on these unstimulated samples is referred to as “direct chromosome analysis.” As maternal cells are not spontaneously dividing in placental biopsies, in addition to the rapid availability of the result, direct chromosome analysis on chorionic villus fragments excludes error introduced by maternal cell contamination. Direct chromosome analysis is not used routinely because the number and quality of complete metaphases suitable for analysis are less consistent than obtained from tissue culture. In addition, the accuracy when compared with cultured preparations is less, in part because of an increased rate of confined placental mosaicism in direct preparations.
Standard laboratory reports will indicate the source of material studied, and the number of cells examined, usually 20, and the type of staining or banding performed. The report may also describe the banding level by subjectively estimating the number of bands in a haploid set of chromosomes. The expected sensitivity of standard G-banding for detection of copy number variants is 5–10 Mb.
The report should provide a karyotype designation, which indicates the number of chromosomes in the cells and the sex chromosome pattern observed. If abnormalities or structural rearrangements are present or when multiple different cell lines are observed, these should be described according to standard karyotype designations described in the International System for Human Cytogenetic Nomenclature (ISCN).
The number of cells examined enables estimation of the confidence intervals for detection of mosaicism at certain percentages of a cell line. See Table 4.1 for estimates of detection of mosaicism by examination of 20 or more cells by diagnostic testing. Additional cells may be examined if there is concern about the initial findings or when there is concern for low-percentage mosaicism or maternal cell contamination.
| No. of cells (n) | Level of Mosaicism Confidence Interval | |
|---|---|---|
| 0.95 | 0.99 | |
| 20 | 14% | 21% |
| 40 | 8% | 11% |
| 50–55 | 6% | 9% |
| 99–112 | 3% | 5% |
| 299–458 | 1% | 2% |
| >459 | 1% | 1% |
If the karyotype is other than normal, a narrative may follow that provides a more understandable description of the findings for the clinician who is not likely to be familiar with complex karyotype nomenclature and abbreviations. The report may also describe clinical implications of unusual or abnormal findings and recommend additional cytogenetic analysis, including testing of the parents, other diagnostic tests, and/or genetic counseling.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here