Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The practice of Pathology Informatics/Clinical Laboratory Informatics is central to all aspects of data stewardship in the clinical laboratory.
Fundamental knowledge of database technology and database principles is critical to understanding key aspects of laboratory information system (LIS) operation.
The LIS is an integral element of the larger enterprise-wide portfolio of information technology solutions that may be rendered as either a stand-alone vendor solution or as an integral module of the larger integrated electronic health record (EHR).
The LIS supports laboratory workflow and automation in all phases of the generation of clinical results (preanalytical, analytical and postanalytical).
Data interoperability is an increasingly important aspect of data stewardship and is supported by a growing ecosystem of standards.
Informatics will increasingly play an important role in data analysis and stewardship as machine learning techniques and solutions are incrementally applied to primary laboratory results, creating derivatized versions of base knowledge, representing incremental and medically actionable knowledge.
Clinical laboratory informatics is a subdiscipline of clinical informatics, which encompasses all of the subject matter of informatics in the general discipline of clinical informatics, and a potpourri of unique topics distinct to the fields of clinical laboratory medicine. This growing assemblage of core material and specialized niche topics poses a challenge for the generalist pathologist or laboratorian to effectively master and utilize in daily practice. Owing to this reality, it is increasingly likely to find pathology groups, both in academia and community practice, making use of specifically trained pathology informaticists who may devote a fraction or even the entirety of their time to this pursuit.
Clinical laboratory informatics encompasses many areas of expertise, with not all of them being within information technology fields per se. The most important aspects of informatics are in many cases nontechnical, with examples being the application of effective interpersonal communications, sound management strategies, robust project management skills, and user experience-centered design. When adding these to the litany of technical subspecialties within pathology informatics, it can be easily seen that the breadth and depth of possible expertise in this area is astonishing, making the case for a multidisciplinary team approach in even midsized projects (noting that it is becoming increasingly unreasonable to expect that any one individual would possess all of the skills and domain expertise typically required for the successful execution of a typical contemporary solution).
Given the significant range of pathology informatics topics and the larger range of clinical informatics topics, this chapter will make selective use of the general framework provided by the American Medical Informatics Association’s “Clinical informatics subspecialty delineation of practice” document ( ), The material contained within this chapter constitutes a reasonable contemporary survey of the areas of expertise and topics that pathology informaticists might typically encounter during their daily activities. In topic areas in which substantial additional domain knowledge is available but exceeds the level of coverage provided by this chapter, appropriate references are provided, along with mention of the significantly expanded knowledge base that can be perused and assimilated as required.
While it is true that clinical informatics—and, specifically, the collective areas of pathology informatics—makes extensive use of information technology, it should be emphasized that these clinical fields are distinct and separate from computer science. They should be viewed from the more holistic perspective of supporting the underlying collective needs of health care in general and the clinical laboratory specifically to generate, analyze, disseminate, and curate primary data. In so doing, raw information is transformed into medically actionable knowledge and wisdom. As such, the practice of clinical informatics and pathology informatics exists at the crossroads of many knowledge domains, including information technology, project management, data sciences and analytics, data curation, change management, and, of course, laboratory medicine. In recognition of this broad scope and vast sets of requisite skills required to practice pathology informatics effectively, it should be immediately seen that to be maximally effective, a team-based, multidisciplinary approach is highly desirable, if not absolutely required. Whereas many, if not most, medical specialties define the value proposition they provide to the overall health care system in terms of direct clinical interaction with patients and procedurally based activities (often carried out in direct contact with the patient), the practice of laboratory medicine fundamentally differs in that the value-added contribution is information itself (and, increasingly, knowledge), in the form of data products that are shared with clinician colleagues and, by proxy, their patients as well. As the information that the clinical laboratory generates is increasingly, if not universally, now in the form of multiple forms of digital communication channels to various downstream information repositories, not the least of which is a health enterprise’s central electronic health record (EHR), it is incumbent on practitioners of laboratory medicine to have definitive command of the constitutive technologies and methodologies that enable effective data curation, transformation, interrogation, and dissemination. This expectation and its associated requirements will only grow in importance as the data that contemporary clinical laboratories now generate increases in both scale and complexity. Most of the core concepts of clinical laboratory informatics covered in this chapter are covered in much greater detail in the Pantanowitz text ( ). Note that anatomic pathology informatics topics as covered in that text are generally beyond the scope of this chapter.
As stated by the Association for Pathology Informatics ( https://www.pathologyinformatics.org/about_api.php ),
“Pathology Informatics involves collecting, examining, reporting, and storing large complex sets of data derived from tests performed in clinical laboratories, anatomic pathology laboratories, or research laboratories to improve patient care and enhance our understanding of disease-related processes. Pathology Informaticians seek to continuously improve existing laboratory information technology and enhance the value of existing laboratory test data, and develop computational algorithms and models aimed at deriving clinical value from new data sources.”
This definition is significant in that it underscores the major role of pathology informatics in transforming raw, laboratory-derived primary data into medically actionable knowledge and, in so doing, creating additional clinical utility. As has been borne out in recent years by the explosive growth of machine learning and general artificial intelligence techniques as applied to laboratory medicine data, it is now well recognized that the primary data generated by the various clinical laboratory sections contains extensive incremental data in encoded form that can be accessed only by advanced analytical and data sciences methodologies. The application of such approaches to primary data as generated by the clinical laboratory is fully within the purview of pathology informatics and its practitioners.
With the advent of commercially available computers, as early as the 1950s, pioneers such as Homer Warner ( ) and Octo Barnett ( ) systematically explored the utility of applying computational and analytical methodologies on health care data sets and workflow models. Up to that point, analysis of such data had been entirely dependent on manual approaches and clerically dependent data entry methods. Besides the obvious benefit of reduction in transcription-based error rates (which can be uniformly seen to be typically between 1.5% and 3%, at minimum), there emerged an expansive range of possibilities for value-added use of primary data once it was properly registered and/or logged in electronic form. In time, this recording process of primary data came to be known as database technology. With its emergence came an expansive portfolio of derivative uses and technologies that continue to expand into the present era.
Commensurate with these early works were attempts to develop both so-called “expert systems” (which were programs that could assist in the rendering of diagnoses and treatment plans, given input data) and EHRs. In their earliest forms, these efforts were only partial successes in terms of both functionality and adoption, underscoring the actual level of complexity inherent in appropriately capturing and curating health information data in surgical/anatomic pathology and laboratory medicine data specifically. Creating applications mirroring actual laboratory workflow has been the critically needed elixir that enabled the development of solutions that truly support productivity and safety. This goal remains one of the singular challenges in the continuous development of new generations of software solutions that support the ever-increasing complexity of the contemporary testing laboratory environment.
No informatics chapter would be complete without at least a cursory introduction and coverage of this important topic, as database technology is a central enabling component of any contemporary laboratory information system. At its simplest possible definition, a database is a program (or assembly of programs, recognizing modern distributed software architecture) that allows for the aggregation, storage, curation, retrieval, transformation, and dissemination of primary data, as well as derivatized and extracted forms of this data. A database serves the very important purpose of adding a critical layer of provenance to every accumulated data element that is housed within it such that both its source and level of confidence concerning its veracity (e.g., patient identity, documentation of results validation prior to release, and history of addended or amended information and/or reports, etc.) can be examined and confirmed upon demand. Such capabilities are central to all of clinical informatics and not just pathology informatics, as information throughout the clinical chart is always subject to change through the accumulation of incremental knowledge as well as by point-wise amendments and corrections to existing records. Just as fundamental as correct initial information and correctly updated records is the need to ensure that all records are updated for correct patient identity, necessitating that positive patient identification be extended to all aspects of laboratory workflow. Ultimately, this has led to the creation of an entire workflow and computational “fabric” of an overall laboratory results verification process so that the correct results are always attributed to the correct patient. Some of the most injurious patient care events have resulted from identification errors (e.g., “wrong blood in tube,” leading to major hemolytic transfusion reactions). This underscores the need for constant vigilance and error proofing in workflow to maintain best practices for validating the identity of all information flowing in or out of the database underlying the laboratory information system.
Recognizing that modern databases (of which the laboratory information is unquestionably a member of this class of information technology solution) are fundamentally transactional systems, it is important to recognize the primary transaction classes that databases facilitate. Known informally as CRUD, short for Create, Read, Update and Delete ( Table 12.1 ), these four basic functions allow for essentially all required information stewardship operations. Some additional commentary on the deletion operation is warranted as, in most cases, actual data deletion is contraindicated for both auditing and patient safety purposes. The highly effective and almost universally implemented alternative approach to data deletion is the use of “inactivation flags,” whereby the database can selectively deprecate data elements and concepts that are no longer in active use, while at the same time keeping them available for any necessary historical look back or auditing activity. Use of this approach is how the important data curation concept of data provenance is maintained. Specific examples of where data deprecation takes place in the laboratory information system include patient name changes and reference interval updates, both of which represent relatively common events. In the case of the latter, for example, a revised reference interval update in the LIS database schema would affect only the reporting of new results. All prior reported results would remain associated with the reference interval with which they were originally associated.
| Function | Definition | Typical SQL Example |
|---|---|---|
| Create | Insert a new table into a database schema or a new data row into an existing table. (Example: inserting a new patient record into the Patient Identity Table) | INSERT INTO Patient_Identity_Table
|
| Read | Extract one or more records from a database. (Example: extracting patient name and DOB when providing a name and medical record number) | SELECT
FROM
Where
and
and
|
| Update | Replace a data element of an existing table row with a new one. (Example: changing the data of birth for a specific patient, using multiple identifiers) | UPDATE
SET
WHERE
and
and
|
| Delete | Remove one or more rows from a table. (Example: remove a patient record row from the Patient Identity Table, using the MRN as the deletion key) | DELETE FROM Patient_Identity_Table WHERE
|
In the language of most database programming (Structured Query Language [SQL]), the four common operations on tables—adding, reading, updating, and deleting (in select circumstances) data—are carried out via four general classes of SQL commands: insert , select , update , and delete . The vast majority of database operations, as carried out on the contemporary lab information system database schema, could be contained within variations of these four general command classes alone. Consequently, having even cursory familiarity with this aspect of the SQL programming language can confer on the laboratorian greatly enhanced ability to converse with vendor database architects, thereby allowing for active participation in any requests for LIS architectural modifications or enhancements. Similarly, for the pathology informaticist, possession of a broader knowledge base for SQL databases and actual SQL programming can be of great utility in efficiently generating custom reports not already provided by the LIS vendor. Such reports can support laboratory operations without the need for additional vendor support, which can be time-consuming and expensive. The availability of such skill sets within the clinical laboratory can be of significant strategic value, recognizing that the generation of specific data extractions for time-sensitive needs (periodic laboratory inspections, state and federal reporting of infectious disease and pandemic data, etc.) is not an infrequent occurrence.
Another key concept integral to database technology and stewardship of any compendium of systematized information is the list of ACID properties: atomicity, consistency, isolation, and durability ( Table 12.2 ). Of these four fundamental concepts, atomicity is perhaps the most important in that it ensures that any intended transaction is either fully executed or not executed at all. To understand the importance of this concept, consider the routine task of updating a patient’s demographics, including name, date of birth, and current address. Typically, such updates are carried out on the core database as a monolithic transaction such that all fields associated with the record in question are updated at the same time. Without the database possessing atomicity as a core property, it would be possible, under certain circumstances, for an update to successfully carry out modification of some fields in a patient’s demographic record, but not all fields. Such a partially completed update would create an errant record, which not only would incorrectly represent the patient, but also could, by coincidence, correctly match a different patient. Such referential integrity problems were manifested in the earliest days of clinical database implementations, and subsequently became far less likely in the setting of adoption of ACID principles by contemporary database solutions.
| Property | Definition |
|---|---|
| Atomicity | Database updates must be all-or-none operations. If a multipart transformation is requested, it must complete in toto or not at all. Thus, the database is left unchanged. |
| Consistency | Change requests to any database table row data must result in the database being left in a form in which all fields are left with valid data. |
| Isolation | Isolation enforces concurrent execution of transactions to operate in the same manner that would have been obtained if the same transactions were executed sequentially. |
| Durability | Engineered software and operating system robustness such that, once a transaction has been committed, it will remain committed even in the setting of system failure (e.g., system crashes, power failures). |
Fundamentally, most contemporary databases are rendered as relational databases, implying that tabular information is stored in separate and distinct tables, which have both rows and columns. By convention, any given database table’s structure defines the columns to represent the distinct classes of information held by a given table and defines rows to represent individual entries housed in a given table, which specify all of the required (or optional) data elements (e.g., the columns) needed to comprise a complete record ( Fig. 12.1 ) for that given table. In addition to externally added content, most database tables will contain one or more keys, which are simply unique identifiers generated by the database itself (typically, sequential numbers or alphanumeric sequences) that serve to uniquely identify each individual row in a table, thereby distinguishing it from all other rows. Keys are also of fundamental importance, as will be subsequently described, in that they form the underlying construct by which a relational database can be realized.
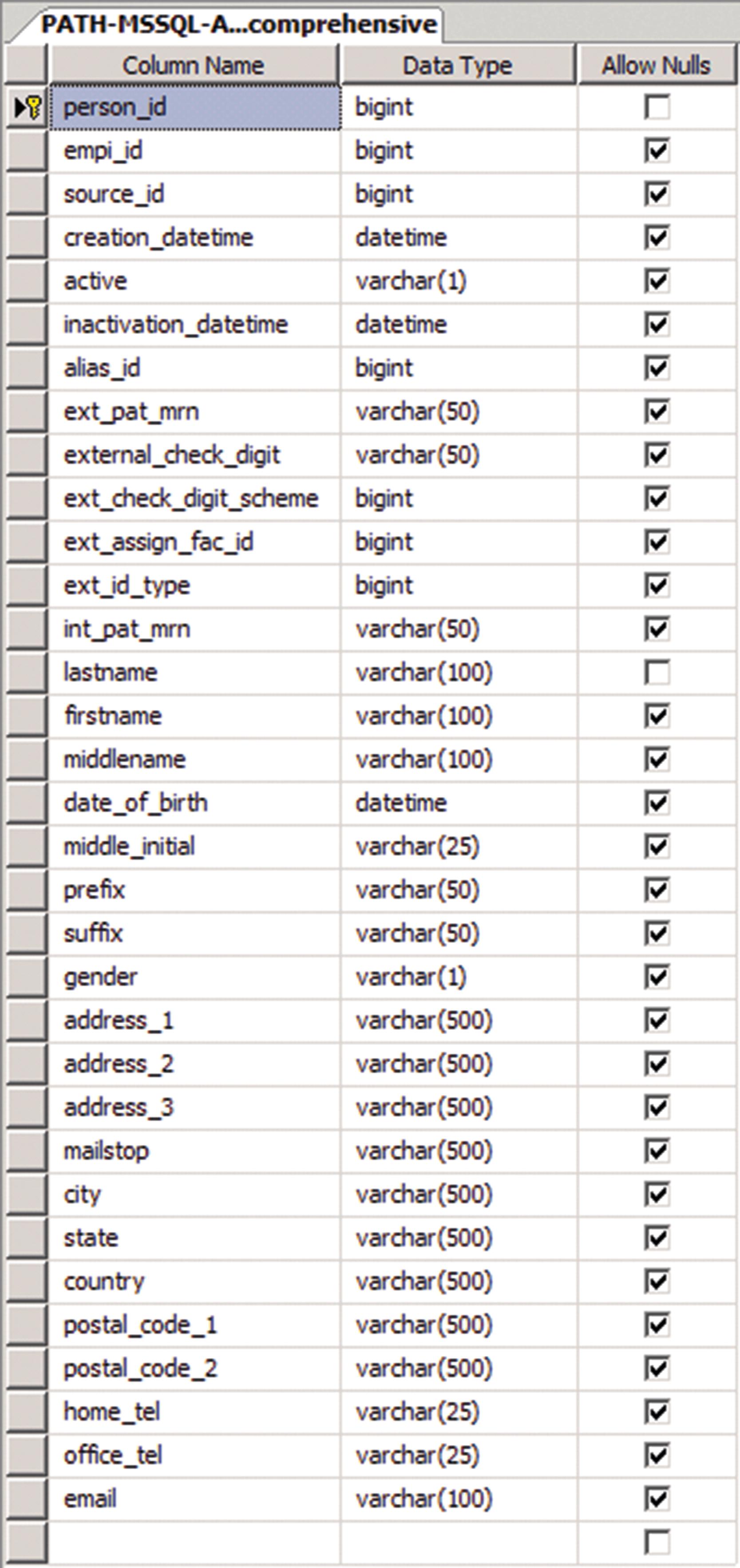
Tables are constructed such that each column can house a particular class of data (e.g., simple text, dates, numbers, etc.) with the option at design time to instruct the database as to whether that class of data will be enforced for that column. With enforcement in place, all attempts to place a nonconforming data element in a constrained column will result in an error. When enforcement is suspended, data classes other than the originally specified type can be included. This type of data storage is defined as a data type overload . There are reasons that a database architect might choose one or more columns to have enforcement of a data class suspended, including the provisioning for general-purpose containers that might need to hold various classes of data concurrently. However, for the vast majority of data representation needs, designing table columns with data enforcement in place is a sound practice, as it ensures that columns with specific intended meaning and function are always populated with appropriate data types and, therefore, correct information.
With the availability of individual tables comes the added potential to link them in meaningful ways such that specific information can be easily recovered while at the same time eliminating (or at least minimizing) the need to reduplicate atomic data elements in multiple tables. As a brief digression, the issue of data reduplication in contemporary database design is a central topic, as any instance in which a unique data element is repeated more than once in the overall database represents an opportunity for the incursion of errors over time. This is especially true as a database grows and evolves. As a simple example, in a hypothetical LIS database in which the system architect chose to include patient demographics in both the orders and results tables, a patient name change requested by the laboratory’s host health organization (a relatively common event) would necessarily require an update in the demographics fields of both tables. As database table count and table complexity would grow, there would be increasing instances in which single data elements are multiply reproduced throughout the overall database. Such growth could represent an exponential (and, thus, unsupportable) increase in complexity, with the associated need to identify and then update all instances of the repeated data element. In the setting of a data element update request, if not all copies of the affected data element are correctly changed to their new correct value, a very real possibility exists for the database to be placed into a condition known as referential integrity failure . For example, a patient’s name would not be represented consistently throughout the multiple tables where it resides, leading to having multiple concurrent names for the same individual in different parts of the overall database schema. Clearly, this is an undesirable outcome.
To address this vulnerability, contemporary relational databases have espoused the notion of data normalization, in which important data elements, such as patient name and medical record number, are contained in only one table of an overall database schema, with all references to such data elements, as may be present in other tables, carried out by use of a pointer to this single referential source data element. This construct, commonly referred to as single source of truth , ensures that when an update takes place to a particular data element, that update is guaranteed to cascade throughout the entire database schema with no possibility that prior values of that data element will remain active.
In specialized settings, in which there is the need for extremely high database performance, it is permissible to reduplicate data elements so that they are immediately available for retrieval without interrogating multiple tables concurrently. Repeating such data elements for performance purposes is known as data denormalization . While very useful in select circumstances, it should be recognized that this process should be carried out with great care to avoid the possibility of referential integrity failure.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here