Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Prenatal genetic carrier screening has evolved over a period of about 25 years. Advances in technology and health policy have ushered this evolution. With these changes, new terminology has appeared, whereas old terms and their definitions have evolved. It is helpful for the reader to understand the current core terminology around the topic of carrier screening.
Condition : There has been a general trend to replace the term “disease” with “condition.” Genetic testing and screening, when applied to populations of healthy people, will commonly uncover health risk to either themselves or their offspring. This can provoke unnecessary anxiety. Furthermore, there is less stigmatization associated with “condition” compared with “disease.”
Primary prevention: A public health strategy aimed at preventing or minimizing the impact on health, such as vaccination in preventing infectious diseases. Prenatal genetic carrier screening can play a role in primary prevention when at risk couples use assisted reproductive technologies (ARTs) in their reproductive planning.
Secondary prevention : A public health strategy aimed at identifying and limiting early disease. Papanicolaou screening for cervical cancer is an example. Carrier genetic screening can play a role in secondary prevention. Postconception decision-making falls into this category of secondary prevention.
Mutation : When referring to changes in the sequence of a gene, this term has generally been replaced by “variant.” Variant has less stigmatization and is less definitive, allowing for classification according to its relationship with a condition of interest (e.g., benign, pathogenic).
Pathogenic variant ( i.e., PV) : A category assigned to variants in a gene that are associated with a condition. The caution here is that penetrance may not be 100%, and there may be variable expressivity (see below).
Likely pathogenic variant : A category assigned to pathogenic variants in a gene where the association with a condition is less certain.
Benign variant : A category assigned to variants in a gene that are clearly not associated with a condition.
Likely benign variant : A category assigned to variants in a gene that are most likely not associated with a condition, but where the benign nature is less certain.
Variant of uncertain significance ( i.e., VUS) : A category of variants in a gene where the variant cannot be placed into either of the above four categories. These variants are numerous and are often the most challenging for clinicians, as they bring uncertainty into management and treatment decisions.
Residual risk : The risk that remains after a negative screening test result. Applied to genetic carrier screening, residual risk is conceptualized by the following formula:
Next-generation sequencing (NGS) : Generally applied to sequencing technologies that allow simultaneous sequence multiple distinct regions of the genome. It is considerably faster and cheaper than traditional Sanger sequencing.
Whole-genome sequencing (WGS): (Now referred to as genome-sequencing by the American College of Medical Genetics and Genomics (ACMG)) Sequencing the entire genome of an individual. This requires NGS.
Whole-exome sequencing (WES): (Now referred to as exome-sequencing by ACMG) Sequencing that selectively targets the protein-coding regions of the genome. Most known pathogenic variants are in these protein-coding regions; therefore, selectively sequencing these regions can reduce cost relative to WGS.
Genotyping: A technological method of assaying genetic variants that targets known pathogenic variants. This approach is faster and less expensive compared with NGS. It allows for simultaneous assay of up to hundreds of thousands of variants. Variants that are not part of the prespecified genotyping platform will not be assayed.
Pan-ethnic screening : This is an approach to carrier screening where all ethnic groups are screened the same way. For example, if pan-ethnic screening for sickle cell disease is the goal, then it is not only performed among individuals of African descent but also applied to all ethnic groups in the same fashion.
Expressivity : Expressivity reflects how a genetic variant can manifest phenotypically. It recognizes that a pathogenic variant can be associated with a range of phenotypes (i.e., variable expressivity). For example, a variant causing Alagille syndrome may present with hepatic dysfunction in some individuals, but not in others.
Penetrance : The percentage of people with a pathogenic variant who develop the associated phenotype. Population-based genotyping has uncovered that many pathogenic alleles are not highly penetrant. Incomplete penetrance makes selecting variants for testing on a carrier screening panel challenging. Incomplete penetrance also challenges interpretation of screening results and downstream clinical decision-making.
Traditionally, carrier screening, as recognized by Wilson and Jungner, was a form of secondary prevention. Its application was largely in public health in the same realm as infectious diseases (e.g., tuberculosis [TB], syphilis). In public health, screening tests are applied to populations broadly (e.g., testing children before enrollment into school) or in a more targeted fashion based on known risk factors (e.g., prisoners). Historically, screening tests were inexpensive and noninvasive. They are generally low technology or amenable to automation, which allows high throughput. Examples include the rapid plasma reagin blood test (RPR) for syphilis or the Mantoux skin test for TB. Because screening tests have an associated false-positive rate, they are often followed with confirmatory diagnostic testing before treatments begin. Diagnostic tests generally have lower false-positive and false-negative rates but can be more expensive and/or more invasive.
Prenatal genetic carrier screening is distinct from primary or secondary prevention screening in several ways that contrast with those described by Wilson and Jungner for traditional screening tests and are important for the clinician to appreciate ( Table 7.1 ):
Targets a broad population (like primary prevention screening).
It is noninvasive (like primary prevention screening).
The same test/method can be used for screening and diagnosis in that there may be no follow-up diagnostic test available or required.
It is generally expensive (like a diagnostic test).
Excluding VUS, the false-positive rate is low (like a diagnostic test).
There is almost always a false-negative rate (e.g., residual risk).
Screening generally has health implications for the offspring and not the person being screened (some exceptions apply).
| Conditions Screened | Condition with a personal/societal burden |
| Well known natural history | |
| Detectable at an early stage | |
| Early stage treatment beneficial | |
| Test Characteristics | Suitable for early stage detection |
| Acceptable by patients | |
| Repeat testing intervals determined | |
| Social | Health care supervision is adequate for increased work burden |
| Physical and psychologic benefits are greater than risks of screening | |
| Costs and benefits are balanced |
Unlike most screening examinations and laboratory tests in which there is a high detection rate and low specificity, prenatal genetic carrier screening generally has high detection rate (DR) for the variants being examined. However, because all variants may not be tested or reported, the false-negative rate can be quite high. This has the potential to yield a low negative predictive value (NPV) compared with what is seen with traditional primary screening, but the low prevalence of specific conditions screened tempers this potential. Residual risk is a surrogate for NPV in clinical genetics practice where carrier screening is the focus. The key components of residual risk are carrier frequency within the population of interest and the screening test DR. There is a residual risk for nearly every condition screened.
These characteristics that distinguish prenatal genetic carrier screening from traditional screening modalities make it a unique entity in public health. These distinguishing features equate to unique challenges around policy, education of patients and providers, as well as implementation.
Unlike screening for one condition at a time (i.e., a la carte testing), expanded carrier screening (ECS) adds breadth (more conditions) and depth (interrogation of or reporting on more variants within a gene). It has a reduced false-negative rate for specific genes (because of the increased number of variants examined) and higher sensitivity for detecting carriers across a broad spectrum of conditions.
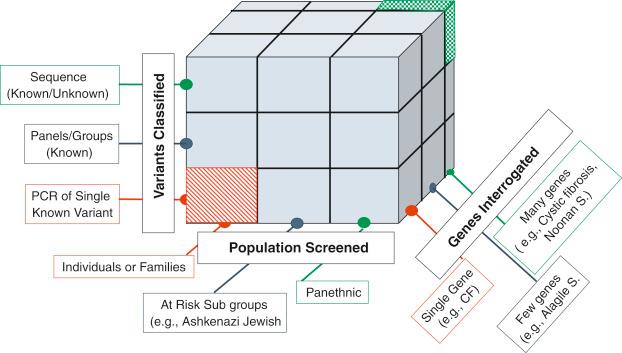
Carrier screening has three dimensions: (1) population(s) screened, (2) gene(s) interrogated, and (3) variant(s) classified. A Rubik cube model illustrates the three axes involved ( Fig. 7.1 ). When prenatal genetic screening first began, small groups of people were the focus (the bottom left cube with red stripes). These small groups included families with an affected proband for a condition or an ethnic group at risk for certain genetic conditions such as individuals of Ashkenazi Jewish descent. Specific conditions were the focus, and a limited number of genetic variants were probed for. With enhanced sequencing technology that included higher throughput, there was an expansion to targeting subpopulations and the number of variants categorized grew. There was no longer an ethnic-based focus on conditions, and screening became less restricted to conditions observed in the family. Over time, screening has allowed for a pan-ethnic paradigm for some conditions.
The human genome comprises approximately 3 billion nucleotides (or letters; A’s, C’s, G’s, and T’s) that serve as the blueprint for all of life. The DNA sequence comprised of these nucleotides, which can be thought of as a string of combinations of the above-mentioned letters/nucleotides, encode for all of the proteins that become part of cells, tissues, and organs. These 3 billion nucleotides are separated into 23 different chromosomes, which are densely packaged segments of DNA. The DNA sequence codes for approximately 20,000 different genes, which are the functional unit of the genome; in general, each gene encodes for a single protein. For example, the CFTR gene encodes for the cystic fibrosis transmembrane conductance regulator protein, a transporter protein that helps transport chloride in and out of cells. However, the fraction of DNA that codes for genes only comprises a small proportion (approximately 1.5%) of the whole genome. Interspersed between the gene “coding” sequences are long stretches of DNA that are “noncoding.” These noncoding regions of the genome have critical regulatory roles in that they determine which genes are turned on in a given cell type or condition. By turning on different genes in different cell types, each cell type can have a specific form and function.
The sequence of everyone’s genome is largely the same. However, the DNA sequence differs at approximately 1 in 1000 nucleotides between any two individuals. These differences in the DNA sequence are called genetic variants. All genetic variants originally arise in a single individual as a mutational event, where the sequence of the genome is copied incorrectly when DNA is passed from one generation to the next. These variants can have a wide range of downstream effects. If the variant occurs in a noncoding region of the genome, it may affect the gene regulatory potential of that DNA sequence, or it may have no effect at all. Similarly, if the variant occurs in a region of the genome that codes for a gene, it may have no effect at all on the resultant protein product, or it may have tremendous deleterious effects on its function, or in some cases even improve the function of that encoded protein.
Genetic variation comes in many forms. The simplest and most common form of variation is single nucleotide polymorphisms, or SNPs, which are single nucleotide changes in the genome. Next are indels, which are short insertions or deletions of several nucleotides in the genome. Lastly, copy number variants (CNVs) and structural variants (SVs) are large structural changes of much greater size (hundreds of nucleotides or more).
The fundamental challenge of diagnostic and prenatal screening is identifying which variants in a gene can cause genetic conditions. Most of the genetic variants in the population are common and thus shared by many individuals in the population. Furthermore, most variants are benign, with no apparent effect on downstream protein function or known condition. Each gene contains many different variants, which have a wide range of effects. There have been considerable coordinated efforts such as ClinGen to distinguish which variants are benign versus pathogenic for disease. It should be noted that the clear majority of variants in a given gene are not pathogenic.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here