Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The kidney plays the primary role in salt and water homeostasis, maintaining osmolarity and volume in the extracellular space within a very narrow range despite wide variations in fluid and salt intake. Aldosterone plays a significant role in the maintenance of mammalian sodium, potassium, water, and acid–base balance, primarily through effects on renal electrolyte excretion. Aldosterone acts through the mineralocorticoid receptor (MR) to promote Na + absorption and potassium, magnesium, and hydrogen secretion by tight epithelia that display high transepithelial electrical resistance and amiloride- and thiazide-sensitive sodium transport. These epithelia are found in distal segments of the nephron, bladder, distal parts of the colon and rectum, ducts of salivary and sweat glands, and choroid plexus. The MR is also extensively expressed in nonelectrolyte transporting cells and is most abundantly expressed in the brain, as well as in other organs including the heart and vessels.
Many of the concepts for sodium transport stimulated by aldosterone were established using amphibian epithelial models such as the toad bladder and the Xenopus laevis kidney A6 cell line. Sodium transport across epithelia is driven by an electrochemical potential difference across the apical membrane allowing for passive movement of ions and water and by an active transport of ions across the basolateral membrane. The apical membrane step is mediated by the opening of the amiloride-sensitive epithelial sodium channels (ENaCs) in the collecting duct and the sodium chloride cotransporter (NCC) in the distal and connecting tubules. The basolateral membrane extrusion of sodium is mediated by activation of the ouabain-sensitive sodium/potassium ATPase.
Steroid hormones bind to and activate intracellular receptors that act as transcription factors that induce the synthesis of specific proteins within the target cells. Early studies demonstrated that aldosterone binds with high affinity to a cytoplasmic and nuclear receptor named the mineralocorticoid receptor in the kidney and other mineralocorticoid target organs, inducing the synthesis of proteins that mediate the best understood effects of aldosterone, sodium retention, and excretion of potassium and hydrogen ions. For many years, despite very early evidence that the heart and vessels were also important direct targets for mineralocorticoid actions, the effects of aldosterone were believed to be limited to the kidney and colon. High-affinity binding sites for aldosterone were also demonstrated in the brain, heart, skin, and other organs. In the early studies, the affinity of the renal receptor for aldosterone was found to be about 40 times greater than that for corticosterone, while the affinity of the receptor in the hippocampus was similar for both corticosteroids. When binding to corticosterone-binding globulin was taken in account, the affinity of aldosterone and corticosterone to the hippocampus and the kidney was similar. Discovery that cortisol and corticosterone were converted to the inactive cortisone or 11-dehydrocorticosterone by the microsomal enzyme 11β-hydroxysteroid dehydrogenase in the vicinity of the MR explained how aldosterone could access the MR in aldosterone target cells despite competition with glucocorticoids that exceed its concentration in the blood by 2–3 orders of magnitude. At the time the concept of prereceptor specificity for steroids was novel, but prereceptor modulation of steroid ligand concentration by specific hydroxylases is now known to occur for most steroid receptors.
The nuclear receptor superfamily is one of the most abundant classes of transcriptional regulators in Metazoans. It comprises a large number of distantly related regulatory proteins that include receptors for hydrophobic molecules such as steroid hormones (estrogens, progesterone, androgens, glucocorticoids, mineralocorticoids, vitamin D, ecdysone, oxysterols, bile acids, etc.), retinoic acids (all- trans and 9- cis isoforms), thyroid hormones, dioxin, sterols, fatty acids, leukotrienes, and prostaglandins. All the members of the steroid/thyroid/retinoic nuclear receptor family of ligand-dependent transcription factors consist of three principal domains: the N-terminal domain (A/B), the DNA-binding domain, and the c-terminal or ligand-binding domain (LBD) ( Fig. 28.1 ).
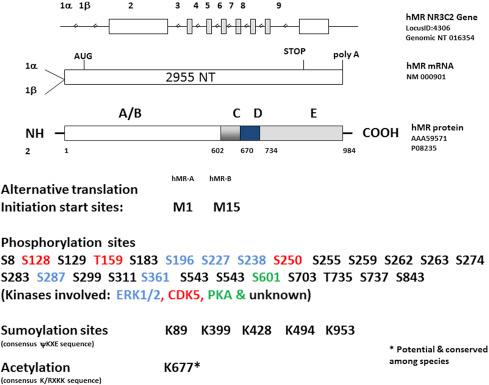
The human MR was cloned by Arriza and Evans and found to encode for a 107-kDa protein with 984 amino acids. Its crystal structure resembles that of other receptors of the same family. The DNA-binding domain of the MR comprises 66 amino acids corresponding to a highly conserved region among members of the nuclear receptor superfamily that has 94% homology with the glucocorticoid receptor (GR) and ∼90% homology with other nuclear receptors. Two groups of four cysteines form α-helices called “zinc fingers,” one of which lies in the major groove of the DNA facilitating specific contacts during transcription. The zinc fingers contain a P box, the interacting surface with the half site of the inverse repeat of the hormone response elements. In the case of the glucocorticoid response elements (GREs), these comprise GGTACAnnnTGTTCT with some variations. The MR uses many, if not all, GREs, but in addition uses specific mineralocorticoid response elements (MREs). The MR elicits MR-specific effects, which include enhanced epidermal growth factor receptor (EGFR) expression by binding to the EGFR promoter within nucleotides −316 to 163 bp of the promoter which does not contain a GRE. The minimal DNA sequence for interaction of the MR and the EGFR promoter is longer than the classical GRE and consists of 15 bp arranged in two 6-bp long inverted repeats separated by 3 bp between them. An additional cotranscription factor, SP1, is required for the MR to activate the EGFR promoter. Activation of this MRE occurs at pathophysiological levels of aldosterone. Intronic MREs outside of the promoter region of the Kcnn4 gene were identified in normal and aldosterone-treated rat colonic epithelial cells in that when sequenced and cloned into a gene reporting system responded only to MR bound to aldosterone, not to MR activated by cortisol. The sequences of these MREs showed low homology to previously described HREs. Chromatin immunoprecipitation using an anti-MR antibody in a differentiated human renal cell line resulted in the identification of 974 genomic MR targets. Computational analysis identified single and multiple half sites and palindromic motifs of MREs of which the most prevalent motif had a sequence of CAgxatGTtCt. Most of these (84%) were located >10 kb from the transcriptional start sites of the target genes. The majority of known MR-binding sites in the genome appear to lack MREs but have recognition motifs for other transcription factors. These findings extend the possibility of new mechanisms by which MR-mediated renal signaling occurs. The DNA-binding domain has an additional nuclear export signal located between the two zinc fingers near the LBD.
The hinge region is located between residues 671–732 and contains a proline stretch, which permits a twist of the DNA-binding domain relative to the LBD, positioning the receptor in contact with the general transcription machinery. This region also possesses a weak ligand-independent nuclear localization signal (NLS1) responsible for receptor subcellular translocation. The hinge region in the GR is potentially responsible for homodimerization, but not heterodimerization; it may play a similar role in the MR.
The crystal structure of the LBD closely resembles that of the glucocorticoid, androgen, and progesterone receptors. The LBD of the human MR has 251 amino acid residues with around 57% homology with the GR and more than 85% homology with MR of other species. It is organized in twelve α-helices and one β-sheet forming three antiparallel layers. The N-terminal domains differ greatly between receptors of the superfamily; the human MR has only 11% homology with the GR; less for others.
The MR has an activating function, AF2, within the LBD that becomes activated in a ligand-dependent manner after agonist binding in the hydrophobic pocket of the LBD. The AF2 is constituted by the H3, H4, H5, and H12 helixes.
The AF2 sequence is highly conserved for all steroid receptor functional domain and is located in H12 of the MR. The positioning of aldosterone is ensured by the hydrophobic residues L938, F941, F946, and F956 of MR helixes H11–12 and is stabilized by the interactions of the 3-ketone, 20-ketone, 21-hydroxyl, and 18-hydroxyl groups of aldosterone to LBD polar residues Q776 and R817, C942, and N770, respectively. The Met852 residue acts as an organizer residue with two major roles. First, it allows steroids with no substituent at the C7 position to be accommodated within the ligand-binding cavity and second, it is involved in the steric hindrance that prevents C7-substituted spirolactones from folding the receptor into its active state. Binding to an agonist results in the rotation of H12, closure of the pocket and rearrangement of helixes H3, H5, and H11, which together expose the outside of the LBD, a hydrophobic groove that interacts with NR box of different coactivators defined by the LXXLL motif (where L is a leucine and X any residue). The L924, Q776R, and L979 mutations in patients with type I pseudohypoaldosteronism impair aldosterone binding and function of the receptor.
The N-terminal domain of the MR, 602 amino acids, is the longest and most highly variable among the steroid receptors. While it has only 15% homology with the N-terminus of the GR, the N-terminus of the MR has been conserved in evolution and is highly homologous among species. This domain contains multiple functional sites responsible for ligand-independent transactivation or transrepression.
Two possible evolutionary events may have led to such a particular intramolecular organization of nuclear receptors. Different domains may have had different origins, with those related to the regulation of metabolism eventually becoming fused to a DNA-binding motif to produce a transcription factor. Alternatively, a multidomain precursor that initially mediated a simple signal transduction mechanism may have acquired increasingly complex functions.
The GR and MR share the highest homology among the members of the steroid receptor family and are derived from a common ancestor. Recent studies suggest that long before aldosterone evolved, the affinity for aldosterone was present in the structure of an ancestral receptor that recognized chemically similar, more ancient ligands. Introducing two amino acid changes into the ancestral sequence recapitulates the evolution of present-day MR specificity. Thus the selectivity of the MR to aldosterone may be more ancient than the hormone itself and that the ancient receptor was activated by a different ligand, perhaps deoxycorticosterone, as is present in certain archaic fish that express MR, but not aldosterone.
On the other hand, the other characters of this plot are the ligands, which share the general property of being small, and hydrophobic molecules derived from common precursors that are also found in plants. Hormonal regulation is dependent neither upon the hormone nor the receptor exclusively, but on both as components of a very complex functional unit , which also includes regulatory proteins including those of the multimeric chaperone system and transcription factors that are cell type and context dependent. In turn, this functional unit is subject to regulation by the differential recruitment of soluble factors, phosphorylation, acetylation, sumoylation, etc. Evolutionary forces probably acted upon the receptor and its ligands as part of the whole functional unit, which is made up of many other proteins that also were evolving.
The human MR gene ( NR3C2 ) spans ∼450 kb in the q31.1 region of chromosome 4 and comprises 10 exons. The first two exons are referred to as exon 1α and 1β and correspond to the 5′ untranslated region in the human. They are followed by eight exons that code for the protein with exon 2 encoding the N-terminal domain (NTD or A/B region). Exons 3 and 4 code for the two zinc fingers of the DNA-binding domain (C region), and the last five exons code for the LBD (E). Rats and mice have an additional 5′ untranslated region. Alternative transcription for these 5′ untranslated exons generates different mRNA isoforms that are differentially expressed in aldosterone target tissues in rats. No evidence exists in humans for a third 5′ UT exon, which may lead to different effects in MR expression and function in humans and common laboratory surrogates when comparative studies are made. MR translation starts 2 bp downstream from the beginning of exon 2; the translated protein is the same for the two (three in the rat) 5′-untranslated isoforms.
Functional splice variants of the MR have been identified. A 12-bp insertion between the two zinc fingers results from the use of a cryptic splice site at the exon 3/intron C splice junction creating a splice variant that is expressed in most tissues with transactivation activities that are not significantly different from the wild type. A 10-bp deletion in the rat and human MR leads to a truncation in the LBD at residue 807 from the rat sequence and is unresponsive to aldosterone. This deletion is expressed at low levels in the rat and human tissues and does not interfere with the wild-type mRNA activity. An additional splice variant skips exons 5 and/or 6, leading to the coexpression of the Δ5 or the Δ5,6 human MR mRNA isoforms. This isoform retains the DNA-binding domain and can act in a weak ligand-independent manner.
The MR has two strong Kozak sequences suggesting the possibility of two different translation initiation sites. While the existence of the two variants has been demonstrated in a transcription–translation type of study, they have yet to be shown to be present in vivo. Use of a monoclonal antibody against amino acids 1–18 of the MR demonstrates that the long variant is expressed, but does not rule out the existence of the shorter one.
The MR is a phosphoprotein with multiple consensus sites for phosphorylation ( Fig. 28.1 ). Rapid phosphorylation of serine and threonine residues occurs within minutes of exposure to aldosterone. These are mediated in part by protein kinase C (PKC) with activation of both PKC alpha and PKC epsilon involved in the rapid, nongenomic effects of aldosterone. There is some evidence that phosphorylation by PKA enhances MR function, but this could be due to phosphorylation of an associated coregulator rather than a direct effect. Inhibition of serine/threonine phosphatases inhibits MR transformation, the conformational changes occurring with ligand binding required for MR binding to DNA. Mutation of one potential tyrosine phosphorylation site, position 73 of the N-terminal domain in the Brown Norway rat (Y73C), was reported to result in robust gain of function or constituent activity of the MR in comparison to the Fisher 344 rat. Position 73 of the MR of the human and mouse are similar to the Fisher 344 MR. However, the original clone obtained from a Sprague Dawley rat also has a tyrosine at position 73 as the Brown Norway rat, and the MR of this strain is not constituently active. Studies with a neuronal cell line have shown that the kinase CDK5 phosphorylates serines 128 and 250 and threonine 159 of the human MR in an aldosterone-dependent fashion. As these phosphorylation sites are within the transactivation domain of the MR N-terminal domain, CDK5 can modulate MR transcriptional activity by affecting its interaction with MR transcriptional cofactors. Roscovitine, an inhibitor of the CDK5, inhibited the aldosterone-induced expression of the aldosterone response gene p75NTR. Phosphoproteomics of the human MR transfected into COS-7 cells demonstrated 16 phosphorylation serines ( Fig. 28.1 ): 14 sites were in the N-terminal domain, 1 each in the hinge region and LBD, and none in the DNA-binding domain. These sites harbor minimal substrate motives for known kinases, except for the serine 843 that matched no known kinase motif. Plasmids encoding the human MR with a phosphomimetic glutamate substitution of serine 843 exhibited no transactivation of a reporter gene containing an HRE, whereas substitution with an alanine had no detectable effect demonstrating that phosphorylation at this site inhibits MR activation. The MR phosphorylated at serine 843 (MR S843-P ) did not translocate to the nucleus when activated by aldosterone. Using a specific antibody that recognized MR S843-P revealed localization to the cytoplasm of renal intercalated cells of mice. Dehydration and angiotensin II infusion resulted in a 50% reduction of the MR S843-P , the effect of angiotensin II was blocked by losartan. Conversely, a high-potassium diet produced a marked increase in MR S843-P 33 . The differential effects of these two different physiological stimuli on dephosphorylation and phosphorylation of the MR is an example of the regulation by different transcriptional and physiological factors on the actions of aldosterone.
Modification by small ubiquitin-related modifier (SUMO) is a posttranslational modification common to most steroid receptors. MR activity can be modulated by sumoylation of both the receptor itself and its coactivator proteins. The MR has four sumoylation consensus motifs in the N-terminal end at positions K89, K399, K428, and one in the LBD at K953 of the human sequence ( Fig. 28.1 ). The consensus motifs for sumoylation are named synergy control motifs and are defined by the consensus ψKXE sequence, where X is any residue and ψ is an aliphatic residue. These sites are highly conserved through evolution. A study of protein–protein interactions resulted in the identification of a member of the protein inhibitor of activated signal transducer and activator of transcription 1 family, PIASxβ, a SUMO E3-ligase that conjugates the MR to SUMO1 in vivo and in vitro , repressing its ligand-dependent transcriptional activity. Transient transfection of the SUMO1-conjugating enzyme Ubc9 increased the ligand-sensitive MR transactivation of reporter constructs using the promoter for the MRE for ENaC, or an MMTV (mouse mammary tumor virus promoter containing a hormone-response element responsive to the MR or GR activation). Mutation of all four lysines of the sumoylation consensus site on the MR eliminated sumoylation by Ubc9; however, transactivation of this mutant MR was still enhanced by Ubc9, indicating that the sumoylation activity is dispensable for the coactivation capacity of Ubc9 and the effects are distinct from the sumoylation of the receptor. Other studies have shown that the transcriptional activity of the MR is modulated by its sumoylation potential, as well as by the sumoylation of MR-interacting proteins. Selective sumoylation of MR interacting proteins may add another dimension to the regulation of MR function.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here