Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
More than 150 years ago, Thomas Hodgkin described several cases of a lymphoproliferative disease, later named Hodgkin disease. For a long time this malignancy, now called Hodgkin lymphoma (HL), has been one of the most enigmatic forms of lymphomas. This is due to several key features of the disease: First, the pathognomonic and suspected tumor cells of HL, the mononuclear Hodgkin and the bi- or multinucleated large Reed-Sternberg cells, are very rare in the tumor tissue, often accounting for only about 1% of the cells. Thus, the molecular analysis of these cells was hampered until methods became available to isolate these cells by microdissection from tissue sections. Second, the Hodgkin and Reed-Sternberg (HRS) cells have a very unusual immunophenotype that does not resemble any normal cell in the hematopoietic system. Therefore, immunophenotyping, which was very informative in revealing the cellular derivation of most other lymphoid malignancies, did not uncover the cellular origin of HRS cells. Third, only a few cell lines could be established from patients with HL, these lines were rather heterogeneous, and for hardly any of these lines, their derivation from the HRS cells in the patient was unequivocally shown. Hence, it was initially difficult to draw firm conclusions from the study of such lines. Although HL still harbors many secrets, exciting novel insights into the cellular origin of the HRS cells and the pathogenetic processes in their generation have been obtained in recent years, which will be discussed in this chapter.
HL is subdivided into classic HL, which accounts for about 95% of cases, and nodular lymphocyte-predominant HL (NLPHL). The tumor cells are called HRS cells in classic HL and lymphocyte predominant (LP) cells in NLPHL (originally the tumor cells in NLPHL were called lymphocytic and histiocytic [L&H] cells ). Classic HL and NLPHL differ in the histologic picture, the morphology and immunophenotype of the tumor cells, and multiple clinical features. Classic HL is further subdivided into four subforms: nodular sclerosis, mixed cellularity, lymphocyte-rich classic, and lymphocyte depletion HL. Again, differences in the histologic picture and HRS cell morphology are the basis for this subtyping (see Chapter 78 , for a detailed description).
Because we now know that HRS and LP cells are derived from B cells (see later), a brief outline of B-cell development and differentiation is given first. B cells are generated in the bone marrow from hematopoietic stem cells in a multistep developmental process. The key determinant for B-cell development is the generation of a functional B-cell receptor (BCR) that is composed of two identical immunoglobulin (Ig) heavy chains and two identical light chains, the latter of which can be of the κ or λ type. B-cell development is initiated when common lymphoid progenitors undergo gene rearrangements at the Ig gene heavy chain locus. The variable part of the antibody heavy chain is composed of three gene segments: variable (V), diversity (D), and joining (J). First, a randomly selected D H gene segment is rearranged to one J H gene segment. In the next step, a V H gene segment is rearranged to a D H -J H joint. A heavy chain can be expressed and the developmental stage of a pre-B cell is reached if the rearrangement is in-frame and productive. In the next step, V κ to J κ rearrangements occur to generate a κ light chain. If this is not successful, V gene rearrangement processes take place at the λ light chain locus. B cells expressing a functional (and nonautoreactive) BCR are released into the periphery and express their BCR as IgM and IgD receptors—that is, two different classes of the heavy chain constant region. The first D H -J H rearrangements are not completely specific for B-lineage cells, but V H D H J H rearrangements and light chain gene rearrangements are highly specific for B cells. Thus, their detection in a cell unequivocally defines that cell as a B cell. Moreover, because of the availability of multiple V, D, and J gene segments and additional diversity generated at the joining sites of the rearranging gene segments, a V(D)J rearrangement (in particular for the heavy chain locus) is unique for each B cell and thus can be used as a clonal marker for B cells deriving from the same mature B cell.
If B cells are activated through the binding of antigen to their BCR and cognate help from T-helper cells, the cells undergo a T-dependent immune response in specific histologic structures, the germinal centers (GC), in lymph nodes or other secondary lymphoid organs. In these follicles, activated B cells undergo massive clonal expansion and further diversify their BCR through two processes: somatic hypermutation and class switching. The process of somatic hypermutation introduces point mutations and some deletions and duplications at a very high rate into the Ig heavy and light chain V region genes. This randomly modifies amino acids in the V regions, and in a selection process involving follicular dendritic cells and follicular T-helper cells, B cells expressing mutated BCR with increased affinity to the stimulating antigen are positively selected, whereas B cells acquiring unfavorable mutations undergo apoptosis within the GC microenvironment ( Fig. 79.1 ). Unfavorable mutations include destructive mutations, such as nonsense mutations, that prevent the expression of a BCR as such. Other disadvantageous replacement mutations still allow BCR expression but reduce affinity to the antigen. After multiple rounds of proliferation, mutation, and selection, positively selected B cells expressing a high-affinity BCR differentiate into long-lived memory B cells or plasma cells and exit the GC. Many GC B cells also undergo class switching before they are selected into the memory or plasma cell pool. In class switching, the originally expressed Cμ and Cδ heavy chain constant region genes (encoding IgM and IgD, respectively) are replaced by downstream located Cγ, Cα, or Cε genes, encoding IgG, IgA, and IgE heavy chains, respectively, so that antibodies with altered effector functions are generated. At all stages of their development, B lineage cells are selected for expression of the appropriate BCR, and cells failing this selection are eliminated.
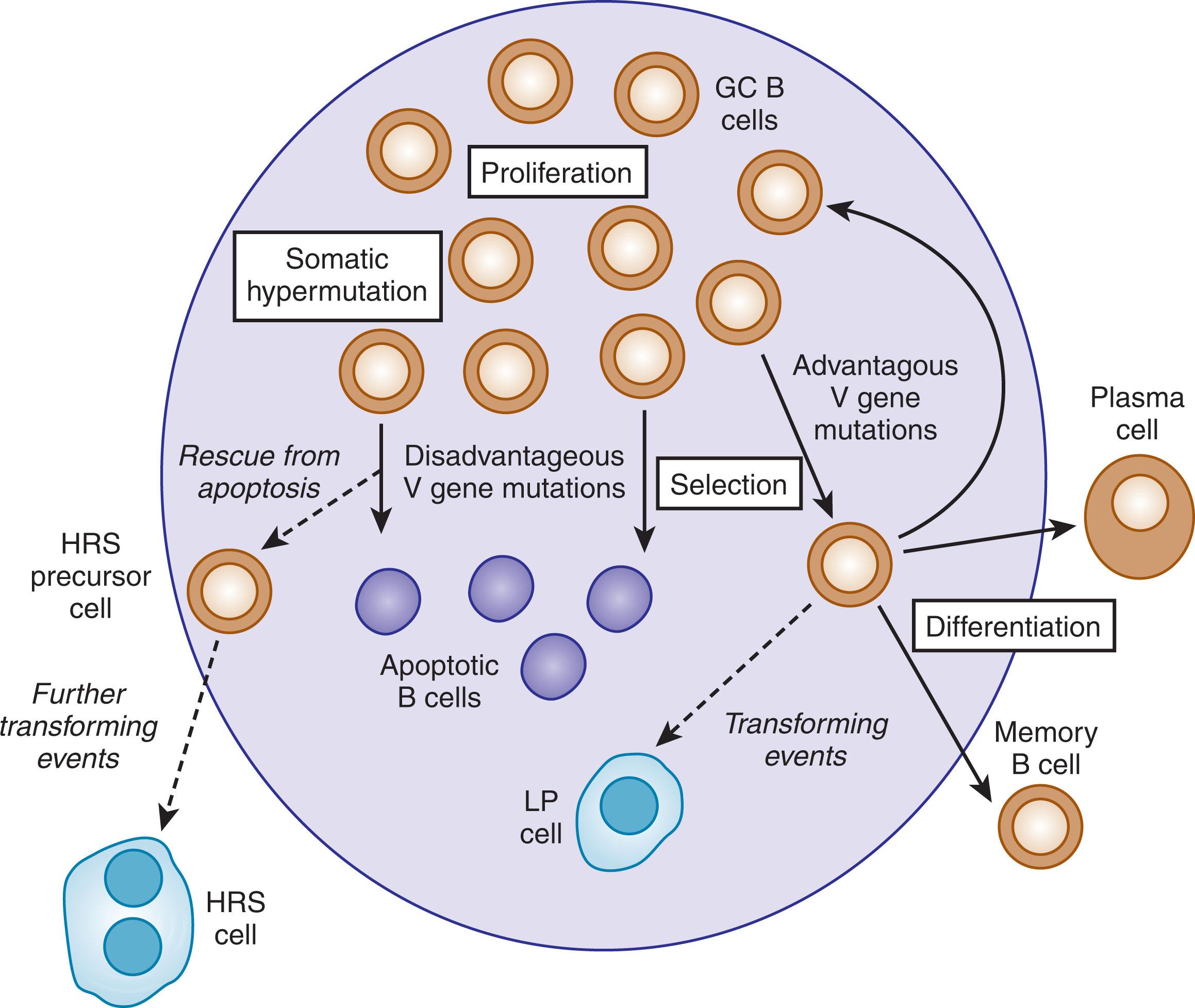
LP cells in NLPHL express multiple typical B cell markers, such as the surface molecules CD20 and CD79 and the transcription factors PAX5, OCT-2, and BOB1 ( Table 79.1 ). LP cells mostly lack expression of markers of other hematopoietic cell lineages, thus their immunophenotype suggests a B-cell origin. Moreover, LP cells are located in follicular structures in close association with follicular dendritic cells and GC-type T-helper cells, suggesting a close relationship with GC B cells. Indeed, LP cells express the transcription factor BCL6, the main regulator of the GC B-cell differentiation program. They also express activation-induced cytidine deaminase (AID), an enzyme that regulates somatic hypermutation and class switching in GC B cells. Thus, all of these phenotypic features point to a GC B cell origin of LP cells (see Table 79.1 ), and this was further corroborated at the genetic level when clonally rearranged and productive Ig V region genes were amplified from isolated LP cells. These rearrangements were always somatically mutated. In some cases, an indication for ongoing somatic hypermutation was observed. Therefore, the Ig V gene analysis further strongly supported a GC B cell origin of LP cells (see Fig. 79.1 ). The detection of the same V gene rearrangements in all LP cells of a given NLPHL case also firmly established for the first time the monoclonality of the LP cells, a hallmark for tumor cells. A gene expression profiling study of LP cells indicated a relationship of these cells to late GC B cells on the way to becoming post-GC memory B cells.
| Feature | Hodgkin and Reed-Sternberg Cells | Lymphocyte Predominant Cells |
|---|---|---|
| Somatically mutated Ig V genes | Yes (very rare exceptions) | Yes |
| Crippling Ig V gene mutations | Yes (>25% of cases) | No |
| Ongoing somatic hypermutation | No | Yes (moderately) |
| Presumed cellular origin | Pre-apoptotic GC B cells | Positively selected, mutating GC B cells |
| Rare cases with a T-cell origin | Yes (<5%) | No |
| B-cell receptor expression | No | Yes |
| Expression of B-cell transcription factors (e.g., OCT2, BOB.1, PU.1, PAX5, E2A) | Rarely and/or at low levels | Yes |
| Expression of B-cell surface antigens (e.g., CD19, CD20) | No or rarely | Yes |
| Expression of GC B-cell markers (e.g., BCL6, AID, HGAL, GCET) | No or rarely | Yes |
| Expression of molecules involved in antigen presentation and interaction with T-helper cells (e.g., CD40, CD80, CD86, MHC class II) | Yes | Yes |
| Expression of markers of non-B cells (e.g., CCL17, NOTCH1, GATA3, ID2, CSFR1) | Yes | No |
| EBV infection of tumor cells | Yes (30%–40%) | No |
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here