Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The discovery of the first mutation in the SCN5A gene encoding the human cardiac voltage-gated sodium channel more than 2 decades ago launched a new era in our understanding of the molecular and genetic basis of arrhythmias. Subsequent work has revealed an unexpectedly large number of clinically and pathophysiologically diverse conditions ( Box 48.1 ) associated with an ever-increasing number of mutations in this gene and in genes encoding important regulatory or interacting proteins. Sodium channel dysfunction or dysregulation caused by gain- or loss-of-function mutations can lead to arrhythmias in the ventricles, atria, or both with potentially life-threatening consequences. The diversity of clinical disorders (phenotypes) associated with SCN5A is explained partly by corresponding heterogeneity in mutant channel dysfunction and resulting cellular electrophysiologic effects. Understanding the functional consequences of SCN5A mutations has driven tremendous progress in elucidating arrhythmia mechanisms in these genetic disorders and has guided new therapeutic strategies.
Congenital long QT syndrome (LQTS) type 3
Brugada syndrome
Idiopathic ventricular fibrillation
Sudden unexplained nocturnal death syndrome
Sudden infant death syndrome
Progressive cardiac conduction system disease
Familial atrioventricular block
Congenital sick sinus syndrome
Atrial standstill
Drug-induced LQTS
Arrhythmia susceptibility in African Americans (S1103Y)
Mixed phenotypes/overlap syndromes
Dilated cardiomyopathy with arrhythmia
Atrial fibrillation
This chapter focuses primarily on pathophysiologic aspects of genetic cardiac sodium channel diseases with an emphasis on the biophysical mechanisms that underlie this channelopathy. The goal of this review is to provide a basic mechanistic foundation for clinical diagnosis, risk stratification, and clinical management. Such mechanistic insights are not possible without a basic appreciation of the channel structure, function, and trafficking as provided in depth by other chapters that are cited throughout this review.
SCN5A encodes the pore-forming subunit of the major voltage-gated sodium channel expressed in the human heart. This gene spans approximately 100 kilobases on the short arm of chromosome 3 (3p21) and consists of 28 canonical exons ranging in size from 53 (exon 24) to 3257 (exon 28) base pairs ( Fig. 48.1A ). The full-length product of SCN5A is a 2016 amino acid protein designated as Na V 1.5, but other nomenclature (i.e., hH1) prevalent in older literature has been used to describe recombinant forms. Native Na V 1.5 is localized mainly to the intercalated discs and, to a lesser extent, to the lateral plasma membranes of cardiomyocytes. Chapter 1, Chapter 9 present the structure and function of sodium channels.
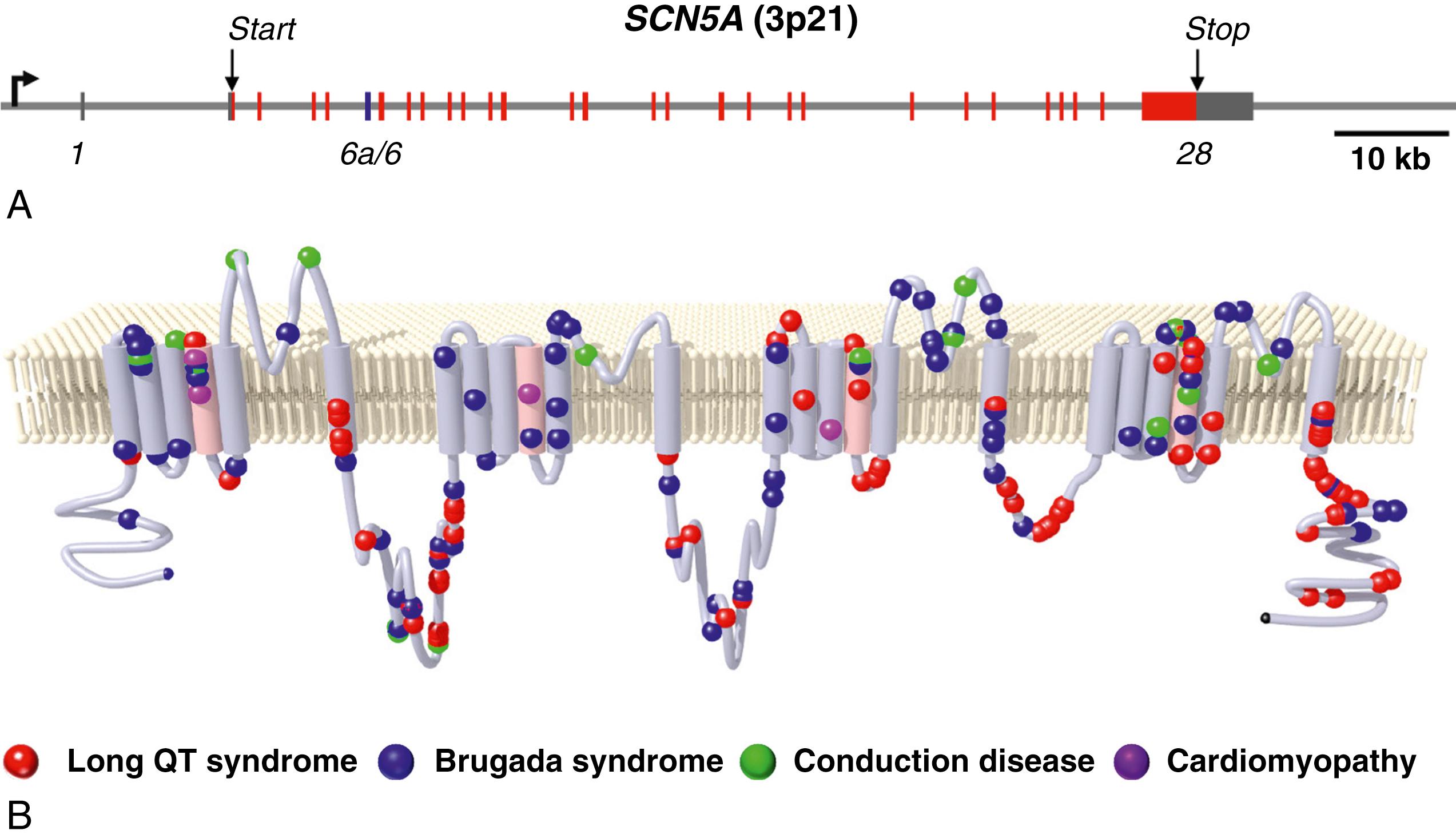
Alternatively spliced messenger RNAs (mRNA) transcribed from SCN5A have been detected in the heart. In fetal and neonatal hearts, another alternatively spliced form of Na V 1.5 is expressed that uses an alternative exon 6 (exon 6a; see Fig. 48.1A ), resulting in several amino acid differences within a voltage-sensor domain (D1/S3-S4). , This fetal-expressed Na V 1.5 splice variant exhibits developmental regulation in the mouse and human hearts and exhibits distinct biophysical properties, including a more depolarized voltage dependence of activation. Other alternative splicing events have less certain physiologic relevance, but certain patterns of alternative splicing involving the carboxyl-terminus arise in heart failure.
Transcription of human SCN5A is under the control of a promoter that precedes the first exon and other determinants of transcriptional activity located in the first intron. Common genomic variants within the SCN5A promoter may influence expression of the gene and have clinical relevance. One particular common variant within an adjacent gene ( SCN10A ) encoding a peripheral nerve–expressed sodium channel (Na V 1.8) has been associated with altered cardiac conduction and Brugada syndrome. , This variant occurs within a binding site for two related T-box transcription factors (TBX3, TBX5) that are involved with control of SCN5A expression through long-range chromatin interactions. , This region of the gene includes an evolutionary conserved cluster of transcriptional enhancer elements that control SCN5A expression.
The cardiac sodium channel resides in the cardiomyocyte plasma membrane as a multiprotein complex consisting of the pore-forming (α)-subunit (Na V 1.5), auxiliary (β)-subunits, and other interacting proteins. A family of four sodium channel β-subunits (β 1 , β 2 , β 3 , β 4 ) encoded by the genes SCN1B, SCN2B, SCN3B, and SCN4B are expressed in the heart and likely interact with Na V 1.5, possibly to modulate channel function or subcellular localization. Several other proteins expressed in the heart also interact directly or indirectly with sodium channels, including ankyrins, caveolin-3, calmodulin, α 1 -syntrophin, glycerol-3-phosphate dehydrogenase 1–like protein (GPD1L), fibroblast growth factor homologous factors, Nedd4-like ubiquitin-protein ligase, multicopy suppressor of gsp1 (MOG1), and 14-3-3η. Mutations in the genes encoding some of these proteins have been implicated in rare cases of inherited arrhythmia susceptibility, suggesting they mediate important functional interactions. Nevertheless, the strength of genetic data supporting the association of interacting proteins with clinical disorders using stringent criteria has been challenged. Further information about regulation of sodium channels in the context of multiprotein complexes is provided in Chapter 18, Chapter 21 .
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here