Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The study of the human cardiac rhythm has long been limited by the study of fundamental mechanisms in the human heart through approximated models of the heart and its environment. None of the common reductionist experimental approaches allows for study of the cardiac system within the natural environment where it functions physiologically. In general, human mechanistic studies are performed in comparable mammalian species for tissue- or organ-level experiments, or in isolated cells. Moreover, in all model systems, genotypic variation cannot reasonably be accounted for because animal models are deliberately homogenized to prevent unpredictable effects caused by genetic variability. Human subject materials are too scarce to allow for meaningful genetic and cause and effect analysis. However, it is becoming increasingly clear that genetic variability may be a key factor in determining the emergence of rare disease phenotypes, both inherited and acquired.
Development of novel technologies has resulted in new ways to study cardiac function and rhythm disorders. One such technology has been induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs). Readily available patient somatic cells, such as fibroblasts, can be reprogrammed to embryonic-like pluripotent stem cells (iPSCs) through application of transcription factors. These iPSCs can further be differentiated to CMs, creating iPSCs-CMs. iPSC-CMs are a patient-derived cell type, thus they retain the donor patient genetic information and response to drugs and disease. The process is depicted in Fig. 9.1 . With the development of iPSC-CMs there is a newfound opportunity to study human CM physiology in vitro, accounting for the effect of human cardiac genetic, cellular, and biochemical characteristics. iPSC-CMs are currently being used in preclinical cardiotoxicity testing and prediction of genotype-phenotype relationships in cardiac disease.
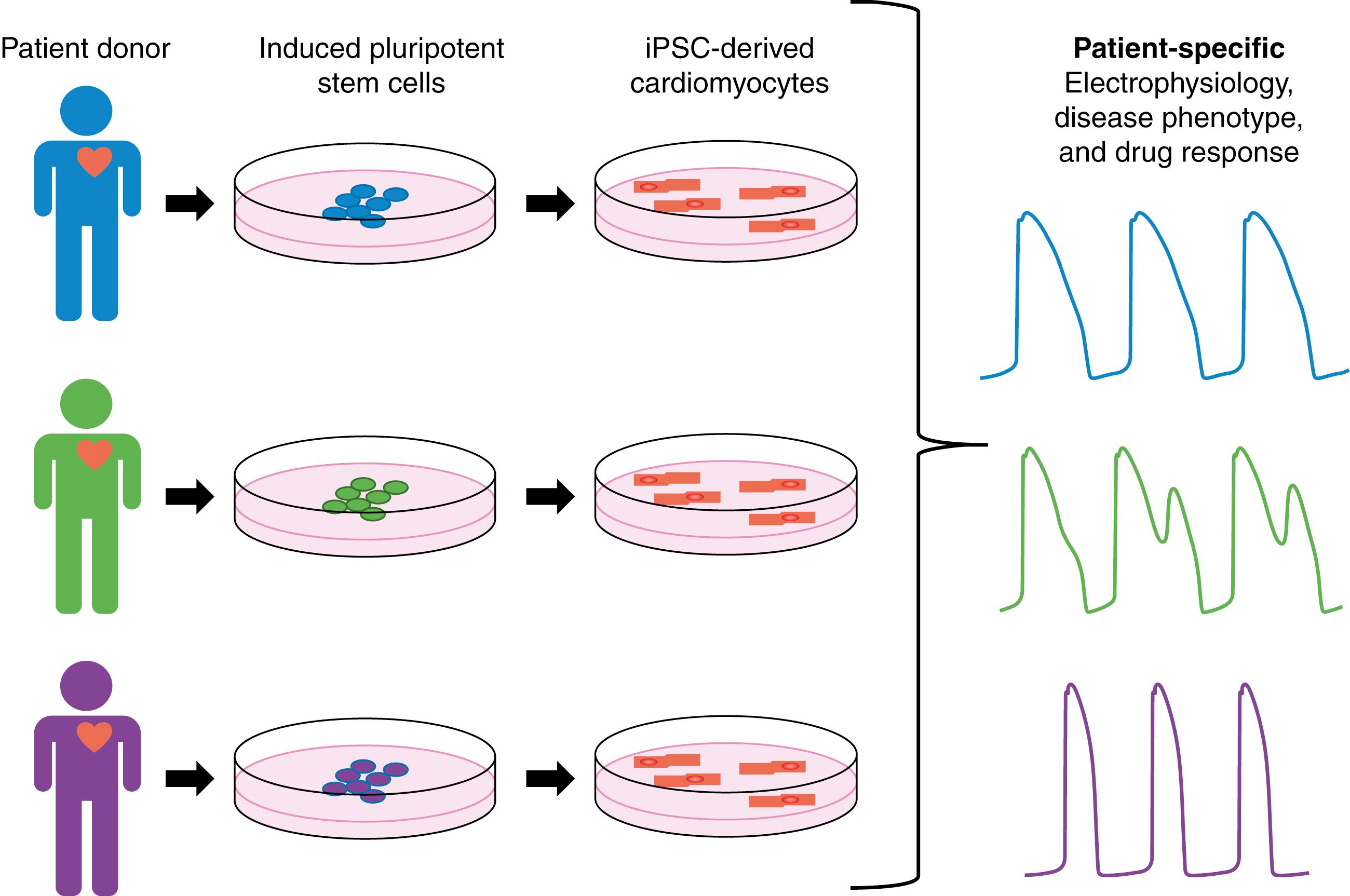
A unique opportunity is now available as iPSC-CM technology allows for the development of patient-specific cellular models of disease. Long QT syndrome (LQTS) is a family of congenital diseases caused by mutations in genes encoding key cardiac ion channels. LQTS can lead to sudden cardiac death in patients and is characterized by a prolongation of the QT interval in the electrocardiogram (ECG), which corresponds to prolonged action potential (AP) duration at the cellular level. LQTS was one of the first diseases studied in iPSC-CMs, as it can be caused by a single identifiable genetic variant, resulting in a change in CM electrophysiology that can be linked to clinical phenotype. In the initial study of LQTS in iPSC-CMs by Moretti and colleagues, iPSC-CMs were created from a donor patient with a mutation in the KCNQ1 gene encoding the potassium delayed rectifier current (I Ks ), known as LQTS type 1 (LQT1). iPSC-CMs from the LQT1 donor patient showed prolonged AP duration compared with control cells. This study also revealed the expected clinical phenotype in response to isoproterenol as iPSC-CMs from the LQT1 patients displayed early afterdepolarizations (EADs) after drug application. Additional studies have also shown patient-specific phenotypes in iPSC-CMs from patients with LQT mutations in I Kr (LQT2) , and I Na (LQT3). , In addition to LQTS, many other cardiac diseases have been modeled in iPSC-CMs, including hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and catecholaminergic polymorphic ventricular tachycardia (CPVT). , , ,
Studying genetic variants within the physiologic context using the iPSC-CM model system provides a platform to interrogate disease mechanisms and study the effects of pharmacology in disease. Underlying mechanisms of disease can be observed within the physiologic context, as opposed to studying ionic current effects in an isolated system such as Chinese hamster ovary (CHO) or human embryonic kidney (HEK) cells. For LQT mutations, studies in iPSC-CMs have revealed disease mechanism related to trafficking defects of hERG and mechanisms of calmodulin mutations on β-adrenergic stimulation and I CaL . Similarly, iPSC-CM studies of HCM and DCM have revealed mechanism of pathogenicity. , Study of disease in iPSC-CMs also provides a method to understand the interaction of pharmacology and disease phenotypes in a physiologically relevant system. For example, treatment with LQT3 patient-derived iPSC-CM with mexiletine has been shown to rescue the LQT3 phenotype, as consistent with the pharmacologic response of the donor patient. In the study of CPVT in iPSC-CMs, patient-derived cells were shown to recapitulate patient-specific responses to pharmacologic intervention, with both clinical and iPSC-CM results showing successful treatment of CPVT with flecainide but unsuccessful treatment with β-blocker therapy.
Although there has been rapid development in the field of patient-specific iPSC-CMs to create models of many cardiac diseases, there are still limitations inherent to the approach. Studies of iPSC-CM disease models have noted cell-to-cell variability within cells derived from a single patient and between studies of the same disease. Moreover, methods to characterize iPSC-CMs are low throughput and limit the ability to leverage the promise of using patient-to-patient variability observed in iPSC-CMs. Patient heterogeneity, as observed in iPSC-CMs, is one of the greatest promises of this technology in studying disease and pharmacologic response. However, without a methodology to understand the sources of variability on a large scale, that promise has not yet been fully realized.
In addition to the study of pharmacologic treatment of genetic disease, iPSC-CMs are also used to predict preclinical cardiotoxicity for a wide range of preclinical and known drugs. Prior to the widespread use of iPSC-CMs, most methods of assessing cardiotoxicity were in mammalian models and did not account for patient-specific genetic heterogeneity. The dominant methods rely on CHO cells, HEK cells, and knockout or overexpression in small rodent models. These experimental methods do not account for the genetic, cellular, or biochemical characteristics of the human heart.
Standard drug screening platforms using single-cell methods primarily examine only the drug effect on hERG potassium channels. Although hERG block is a common surrogate indicator for susceptibility to arrhythmia, it has also been shown to be an incomplete and flawed arrhythmia marker. The exclusive use of hERG as a surrogate indicator for arrhythmia has likely prevented a number of effective drugs from reaching the marketplace with low risk for proarrhythmia. Even in later preclinical stages of drug development, testing in animal models is not always an accurate representation of the human physiologic environment. In particular, animal models cannot recapitulate patient-to-patient variability, which often leads to adverse drug effects. Thus the existing methods for pharmacology testing limit the feasibility of drawing conclusions on human safety or therapeutic effects.
One critical advantage of iPSC-CM-based drug testing is in the patient-specific nature of iPSC-CMs. Drug response is widely known to vary among human subjects. This includes subjects with variable vulnerability to the cardiotoxic effects of certain drugs. Notably, this phenomenon can be recapitulated in patient-specific iPSC-CMs. Several studies have shown that patient-derived iPSC-CMs recapitulate patient-specific drug responses; AP parameters in patient-derived iPSC-CMs recapitulate clinical QT prolongation in healthy donor patients. , At least one study has shown that variability in iPSC-CM drug responses may limit the current applicability of these patient-specific approaches, in part because of small sample sizes.
However, in some applications, it is clear that iPSC-CMs can provide valuable prediction of off-target cardiotoxic effects. Improving understanding of unintended cardiotoxic effects may allow more useful drugs to make it to market. In one study of patient-specific cardiotoxicity of breast cancer patients to doxorubicin, iPSC-CMs from patients with drug-induced cardiomyopathy recapitulated a reduced viability after drug application. Importantly, iPSC-CMs from patients without doxorubicin-induced cardiotoxicity recapitulated the noncardiotoxic cellular phenotype. In a study by Sharma and colleagues, iPSC-CMs were tested with a panel of anticancer drugs, allowing for the creation of a cardiotoxicity score based on iPSC-CM response to drugs and known clinical cardiotoxic effects. Several other studies have also worked to develop methods of testing cardiotoxicity using iPSC-CMs. , In addition, cardiotoxicity testing in the physiologic context of iPSC-CMs allows for the development of multitherapy approaches to combat off-target cardiotoxic effects. ,
In July 2013, the US Food and Drug Administration (FDA) and other partners developed the Comprehensive In Vitro Proarrhythmia Assay (CiPA). They outlined a two-pronged nonclinical approach to drug cardiotoxicity testing. The first prong of CiPA is the development of a computational model to predict the effect of drug interaction with ion channels in cardiac cells. The second prong is the performance of electrophysiologic tests on stem cell–derived CMs to validate the predictions of the computational model. Several studies have shown that iPSC-CMs accurately recapitulate clinical expected effects of proarrhythmic drugs. , CiPA has only recently become a realistic alternative to current preclinical protocols because of advancements in the technology to differentiate CMs from pluripotent stem cells and improvement of in silico models for adult CMs. CiPA is widely regarded as a revolution in cardiotoxicity testing, but it is just the beginning. An approach combining stem cells and in silico testing may lead to an entirely new era in preclinical drug screening for efficacy and toxicity.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here