Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
It is widely recognized that heritable genetic variation (i.e., genotypes or haplotypes) can translate into inherited phenotypes, some of which predispose to or cause diseases and others alter response to treatment. One aim of medical genetics and pharmacogenomics (PGx) is to understand the myriad associations between inherited genotypes and specific phenotypes of disease or drug response, with the ultimate goal of better defining the risk for, or outcome of, diseases and the response to specific medications. In cancer, disease prognosis and treatment response can be affected by both inherited (germline) and acquired (somatic) genome variation, and both types of genome variation have been shown to alter the effects of certain medications. Many seminal discoveries in medical genetics were made in the course of investigating hematologic disorders, with hemoglobinopathies among the most prevalent monogenic disorders, affecting approximately 7% of the world's population. PGx also has a long tradition in hematology; one of the first documented clinical observations of inherited differences in drug effects was the relationship between hemolysis after antimalarial therapy and the inherited glucose-6-phosphate dehydrogenase (G6PD) activity in erythrocytes.
In the pregenomic era, efforts concentrated on mapping highly penetrant monogenic (mendelian) loci for both specific diseases and drug-metabolizing pathways that influence the effects of medications. Completion of the Human Genome Project and the development of arrays for genome-wide single-nucleotide polymorphism (SNP) and DNA methylation analyses, “next-generation” DNA (whole-exome sequencing [WES; coding regions only] and whole-genome sequencing [WGS; coding and noncoding regions]), and RNA sequencing (RNA-seq) technologies have enabled relatively inexpensive and essentially agnostic genome-wide approaches to identify genomic variants that predispose to diseases and/or modify drug responses and/or contribute to heterogeneity of monogenetic disorders and complex diseases that are polygenetic in nature. More recently, single cell sequencing has allowed further resolution of the heterogeneity in genome variation in diseases such as hematologic malignancies. In addition to genome sequence variation, epigenetic differences are increasingly recognized as important for the development of diseases and contribute to differences in the pharmacologic effects of many medications, referred to as pharmacoepigenomics . This chapter provides a brief overview of PGx and pharmacoepigenomics, using selected examples to illustrate its current and potential impact on the treatment of hematologic diseases.
The genome-wide systematic identification of heritable (i.e., germline) and acquired (i.e., somatic) variants, and the functional analysis of genes, their variants, their expression, and their related products (i.e., proteins) have revolutionized the study of many diseases, the development of new medications, and the optimization of drug therapy. Genomics, transcriptomics, methylomics, and metabolomics increasingly enable more comprehensive assessments of a person's risk for acquiring a particular disease, to identify drug targets, and to explain interindividual differences in the effectiveness and toxicity of medications.
The Human Genome Project and subsequent projects such as the International Genome Sample Resource (IGSR) and 1000 Genomes Project (1KGP), and the WGS/Trans-Omics-Precision Medicine (TOPMed) have unveiled many types of variations within the 3.27 billion base pairs (bp) of the human haploid genome (Genome Reference Consortium Human Build 38 patch release 13 [GRCh38.p13]) ( Table 8.1 ); the spectrum ranges from single base-pair differences to large chromosome events, such as insertions or deletions of chromosomal DNA, or structural variants (translocations and other genomic rearrangements that affect more than 50 bp of sequence). For practical purposes, the term sequence variation is mainly used herein. Polymorphisms are defined as common inherited variations in DNA sequence that are typically, although somewhat arbitrarily, defined as the least common allele having a frequency of 1% or more in the population.
| Genomic Variants | Description | Address |
|---|---|---|
| National Human Genome Research Institute (NHGRI) | Website of the NHGRI with the aim to improve human health by genome research | http://www.genome.gov/ |
| GenBank | NIH genetic sequence database—an annotated collection of all publicly available DNA sequences | http://www.ncbi.nlm.nih.gov/genbank/ |
| Genome Reference Consortium (GRC) | Produce a single consensus representation of the human genome | https://www.ncbi.nlm.nih.gov/grc/human |
| The International Genome Sample Resource (IGSR) and 1000 Genomes Project (1KGP) | A deep catalog of human genetic variation | https://www.internationalgenome.org/ |
| Database of Short Genomic Variations (dbSNP) | Repository of all types of short genetic variations <50 bp in length | http://www.ncbi.nlm.nih.gov/SNP/ |
| Database of Human Genomic Structural Variation (dbVar) | Archive of large-scale genomic variants (generally >50 bp) such as insertions, deletions, translocations and inversions | http://www.ncbi.nlm.nih.gov/dbvar/ |
| ClinVar | Archive of reports of the relationships among human variations and phenotypes | http://www.ncbi.nlm.nih.gov/clinvar/ |
| Database of Genomic Variants (DGV) | Catalog of human genomic structural variation | http://dgv.tcag.ca/dgv/app/home |
| Encyclopedia of DNA Elements (ENCODE) | Project to identify all functional elements in the human genome sequence | http://www.genome.gov/10005107 |
| Roadmap Epigenomics Project | Public resource of human epigenomic data | http://www.roadmapepigenomics.org/ |
| GWAS Catalog NHGRI-EBI | Catalog of human GWASs | https://www.ebi.ac.uk/gwas/ |
| Pediatric Cancer Genome Project | Decoded the complete and normal genomes of ~800 pediatric cancer patients | http://www.pediatriccancergenomeproject.org/site/ |
| TOPmed Program | Short-read WGS data from 53,831 individuals | https://www.nhlbiwgs.org/ |
| Pharmacology and PGx | Description | Address |
|---|---|---|
| Pharmacogenomics (PGx) Knowledge Base | Most comprehensive website on PGx | http://pharmgkb.org |
| Clinical Pharmacogenetics Implementation Consortium (CPIC) | Provides guidelines that enable the translation of genetic laboratory test results into actionable prescribing decisions for specific drugs (see Table 8.2 ) | https://www.pharmgkb.org/page/cpic |
| US Food and Drug Administration (FDA)—PGx Biomarkers | Contains a list of FDA-approved drugs with PGx information in their labeling | http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm |
| The miRNA PGx Database | Identifies associations of miRNAs, genes they regulate, and the drugs annotated in literature as dependent on these genes. | http://www.pharmaco-mir.org/about |
| Pharmacogene Variation Consortium (PharmVar) | Repository for pharmacogene variation that focuses on haplotype structure and allelic variation | https://www.pharmvar.org/ |
| PGRN-RIKEN | Data from published GWAS of PGx research | https://www.pgrn.org/riken-gwas-statistics.html |
| Genetic Testing Registry (GTR) | Central location for voluntary submission of genetic test information by providers | https://www.ncbi.nlm.nih.gov/gtr/ |
| The UCSF-FDA TransPortal | A public drug transporter database | http://dbts.ucsf.edu/fdatransportal/ |
| The IUPHAR/BPS Guide to PHARMACOLOGY | Database with quantitative information on drug targets and their ligands | https://www.guidetopharmacology.org/about.jsp |
| Clinical Interpretation of Variants in Cancer (CIViC) | Community-driven web resource for clinical interpretation of variants in cancer | https://civicdb.org/home |
The most common and important inherited sequence variations are SNPs, positions in the genome where individuals have inherited a nucleotide that differs from the most common sequence (“wild-type”) at the position in the genome. Many efforts are underway to catalog these variants, because a comprehensive SNP catalog offers the possibility to pinpoint important variants in which nucleotide changes alter the function or expression of a gene that influences diseases or response to medications. The main public database is the “Database of Short Genetic Variations” (dbSNP; a repository of genetic variations less than 50 bp in length) and a growing number of SNPs (currently approximately 135 million validated; dbSNP Build 150) has recently been driven largely by the 1KGP and TOPMed project (see Table 8.1 ).
SNPs are present throughout the genome, in exons, introns, promoters, enhancers, and intergenic regions. To elucidate the relationship between SNPs and phenotypes of interest, initial efforts have concentrated mainly on SNPs that are likely to alter the function or expression of a gene. However, only a small portion of the identified SNPs lies within coding regions; only about half of those SNPs cause amino acid changes in expressed proteins, and only a subset of those alters the function of the encoded protein (“damaging SNPs”). SNPs that cause amino acid changes are referred to as nonsynonymous SNPs (nsSNPs) and are the main sequence variants underlying most of the highly penetrant inherited monogenic diseases currently known, such as hemoglobinopathies. The likelihood that nsSNPs will result in disease or functional changes in drug metabolism or transport depends on the localization and nature of the amino acid change within the encoded protein; multiple software algorithms have been developed to “predict” whether a certain amino acid change is likely to have a major or minor effect on protein function (i.e., “damaging” versus “nondamaging”).
Although it is intuitively obvious that amino acid substitutions have the potential to change the function of a protein, gene expression also can be affected by SNPs positioned in regulatory sequences or intronic regions. For example, a “silent” or synonymous SNP has been identified that affects protein folding and function of an important drug transporter, namely adenosine triphosphate (ATP)–binding cassette (ABC) transporter ABCB1, and this variant has the potential to influence the intracellular accumulation of drugs that are substrates for ABCB1 (transports some drugs out of cells). Moreover, SNPs in the promoter region can alter the regulatory promoter function and the gene's expression, thereby influencing drug effects. Using a genome-wide association study (GWAS), children and adults who were homozygous for the rs924607 T allele polymorphism in the promoter region of the gene encoding a centrosomal protein involved in microtubule formation (CEP72) were found to be significantly predisposed to vincristine-induced peripheral neuropathy, and in vitro experiments have shown that the CEP72 promoter rs924607 T allele creates a binding site for a transcriptional repressor leading to lower expression of CEP72 messenger RNA (mRNA) and increased sensitivity of neurons and acute lymphoblastic leukemia (ALL) cells to vincristine.
In addition, diverse classes of small to long noncoding RNAs (ncRNAs) have emerged as important regulators of gene expression and genome stability. For example, micro-RNAs (miRNAs) are small (19 to 22 nucleotides), single-stranded RNA molecules that can influence cellular mRNA levels or impair translation after binding to miRNA binding sites at the target gene’s 3′-untranslated region. SNPs in miRNA binding sites or in the sequence encoding miRNAs have the potential to alter binding and function of miRNAs, respectively. Indeed, a so-called miRSNP, which is defined as a functional SNP that can interfere with miRNA function, had been reported to affect the expression of the antifolate target dihydrofolate reductase, thereby influencing antifolate pharmacodynamics. The miRNA PGx Database (Pharmaco-miR) is a helpful research resource to identify associations of miRNAs, genes they regulate, and the drugs annotated as being dependent on these genes (see Table 8.1 ).
Collectively, these examples demonstrate that SNPs in functionally different genomic regions can influence drug disposition and response.
Structural variations are balanced or unbalanced changes in DNA content and encompass alterations ranging from submicroscopic sequence variants greater than 50 bp to larger, sometimes cytogenetically visible, variants. Unbalanced DNA alterations that change the number of base pairs in comparison with a reference genome are as frequent as or even more common than SNPs and include copy number variants (CNVs) or smaller insertions/deletions (indels). Balanced variations such as inversions and translocations are less common in germline DNA but are often found as somatic genome variants in hematologic malignancies (e.g., ALL). Many efforts focus on the identification, validation, and mapping of these variants, and the major catalogs are the Database of Genomic Variants (DGV) and the Database of Human Genomic Structural Variation (dbVAR; see Table 8.1 ). CNVs are found in a wide spectrum of genomic regions; therefore many pharmacologically relevant genes can be affected by these variants. Indeed, CNVs have been described to influence activity of some of the most important drug-metabolizing enzymes, such as cytochrome P450 enzymes and glutathione S -transferases.
Genomic instability is a hallmark of cancer cells. Nonrandom genetic abnormalities, including aneuploidy (gains and losses of whole chromosomes) and structural rearrangements that often result in the expression of chimeric fusion genes (e.g., BCR-ABL1 ), can be found in the majority of hematologic malignancies. These acquired (somatic) genomic variations can differ significantly from inherited (germline) genomic variations and can, for example, create allele-specific copy number differences between normal host cells and cancer cells. Such differences can have pharmacologically relevant consequences. Indeed, it was shown that the cellular acquisition of additional chromosomes in leukemia cells—for example, the gain of additional chromosomes 21 in hyperdiploid ALL (more than 50 chromosomes)—can cause discordance between germline genotypes and leukemia cell phenotypes, which are important when these discordant genotypes/phenotypes influence the disposition of antileukemic agents. Moreover, somatic deletions of genes encoding proteins that regulate the stability of the DNA mismatch repair enzyme mutS homolog 2 (MSH2) have been identified in approximately 11% of children with newly diagnosed ALL. These deletions in ALL cells have been shown to cause DNA mismatch repair deficiency and increased resistance to thiopurines (TPs), representing another genomic mechanism by which leukemia cells can acquire MSH2 deficiency and mercaptopurine (MP) resistance. In addition, recent comprehensive genomic analyses comparing paired diagnostic and relapsed ALL samples provide evidence for a darwinian selection process of relapse-associated somatic mutations in genes encoding key enzymes involved in antileukemic agent activation/inactivation (e.g., TP resistance due to variants in genes involved in TP inactivation [e.g., NT5C2 or PRPS1 ], methotrexate [MTX] resistance due to variants in FPGS which encodes the cellular MTX activating enzyme folyl-polyglutamyl synthetase, and glucocorticoid (GC) resistance due to variants in the glucocorticoid receptors (GCRs) NR3C1/2 , and variants in genes encoding epigenetic regulators which affect GC response [e.g., CREBBP , and WHSC1 ]) during disease progression.
Cataloging the pattern of genome variation in diverse populations is fundamental in understanding areas of human phenotypic diversity such as interindividual and interethnic differences in drug responses; increasingly detailed maps of human genomic variation are provided in public databases (see Table 8.1 ). Information from these maps has been used to design high-throughput genotyping platforms (e.g., SNP chips), thereby providing tools to interrogate the relationship between genetic variation across the human genome and important phenotypes such as disease or response to medications in a relatively unbiased (agnostic) fashion.
SNP catalogs have been used in GWASs to pinpoint genes important to diseases and drug responses, and approximately 10% of published GWAS have focused on PGx. International consortia like the Pharmacogenomics Research Network–RIKEN Global Alliance (PGRN-RIKEN) were established to facilitate GWAS PGx studies, and a publicly available resource—the National Human Genome Research Institute–European Bioinformatics Institute (NHGRI-EBI) GWAS Catalog—provides an updated summary of published GWASs (see Table 8.1 ).
Epigenetics encompasses inherited and acquired changes in gene function that cannot be explained by alterations in sequence of nucleic acids. The epigenome is a complex layer of regulatory information that is superimposed on the genome ( epigenetics literally means “above genetics”), with major mechanisms that contribute to epigenetic variation including DNA methylation, DNA hydroxymethylation, and various histone modifications such as histone acetylation and methylation. As in medical genetics, many seminal discoveries in medical epigenetics were made during investigations of hematologic diseases, and the myelodysplastic syndrome is considered a prototypical example of an epigenetic disease. In contrast to stable sequence variants, the epigenetic cellular state is principally malleable and can be influenced by environmental factors such as diet and toxin exposure. Of note, the expression of genes that encode important drug-metabolizing enzymes (e.g., cytochrome P450) and drug transporters (e.g., solute carrier [SLC] family) have been shown to be altered via intrinsic and extrinsic factors that modify the epigenetic signature, thereby influencing the disposition and effects of drugs. Moreover, the dynamic nature of epigenetics provides a mechanism to modulate the expression of genes that influence drug sensitivity, and so-called epidrugs (i.e., drugs that influence gene expression via epigenetic mechanisms) have already been successfully incorporated into the treatment of hematologic diseases (e.g., hypomethylating agents such as decitabine and azacitidine for myelodysplastic syndrome, and histone deacetylase inhibitors (HDACis) such as vorinostat for acute myeloid leukemia [AML], belinostat for T-cell lymphoma and romidepsin for cutaneous T-cell lymphomas).
Efforts are ongoing to generate detailed epigenomic maps to provide a basis for understanding cellular processes, the pathogenesis of diseases, and alterations in drug responses, such as the Encyclopedia of DNA Elements (ENCODE) and the Epigenomics Roadmap (see Table 8.1 ).
PGx is a major element of the Precision Medicine Initiative announced by US President Barak Obama in 2015. Currently, empiric approaches are typically used to select drug therapy for most patients and most diseases, despite the fact that there is great heterogeneity in the way people respond to medications, in terms of both host toxicity and treatment efficacy. Unfortunately, for almost all medications, interindividual differences are the rule, not the exception, and these differences result from the interplay of many variables, including genetics and environment. Variables influencing drug response include pathogenesis and severity of the underlying disease being treated; drug interactions; the patient's age, sex, nutritional status, and renal and liver function; the presence of concomitant illnesses; and other components of treatment. In addition to these clinical variables, both inherited and acquired (e.g., somatic mutations in cancers) genome variation can influence the disposition and effects of medications, including many used to treat hematologic diseases. Clinical observations of inherited differences in drug effects (based on family studies and twin studies) were first documented in the 1950s, and the concept of pharmacogenetics was defined initially in 1959 by Friedrich Vogel as “the study of the role of genetics in drug response.” The number of recognized clinically important pharmacogenetic traits grew steadily in the 1970s; the elucidation of the molecular genetics underlying these traits began in the late 1980s and 1990s, with their translation to molecular diagnostics to guide drug therapy being well underway in the 2000s. The study of pharmacogenetics began with the analysis of genetic variations in drug-metabolizing enzymes and how those variations translate into inherited differences in drug effects. Subsequently, the field has incorporated genome-wide approaches to identify networks of genes that govern the clinical response to drug therapy (i.e., PGx). However, the terms pharmacogenetics and PGx are generally considered to be synonymous for all practical purposes. With the recognition that epigenetic modification affects gene expression and can contribute to variability in drug effects, the field of pharmacoepigenomics has gained additional attention and importance.
Overall, PGx can be viewed as a broad strategy to establish models of drug disposition and effects by integrating information from genome sequencing, functional genomics, high-throughput molecular analyses, pharmacokinetics (e.g., drug metabolism and disposition), and pharmacodynamics (treatment response, adverse drug reactions [ADRs]). Approaches to establish PGx models include candidate gene analyses (which focus on the analysis of single genes or sets of functionally related genes in pathways thought to be important for the medicine under study) and more agnostic genome-wide analyses. PGx models can be used to maximize efficacy and reduce toxicity of existing medications and to identify novel therapeutic targets.
Comprehensive reviews on PGx and pharmacoepigenomics are available elsewhere. Herein, clinically relevant examples are provided to illustrate the potential of PGx and pharmacoepigenomics to improve current drug therapy for hematologic disorders, to prevent hematologic toxicity, and perhaps to identify novel targets for developing new therapeutic approaches in hematology.
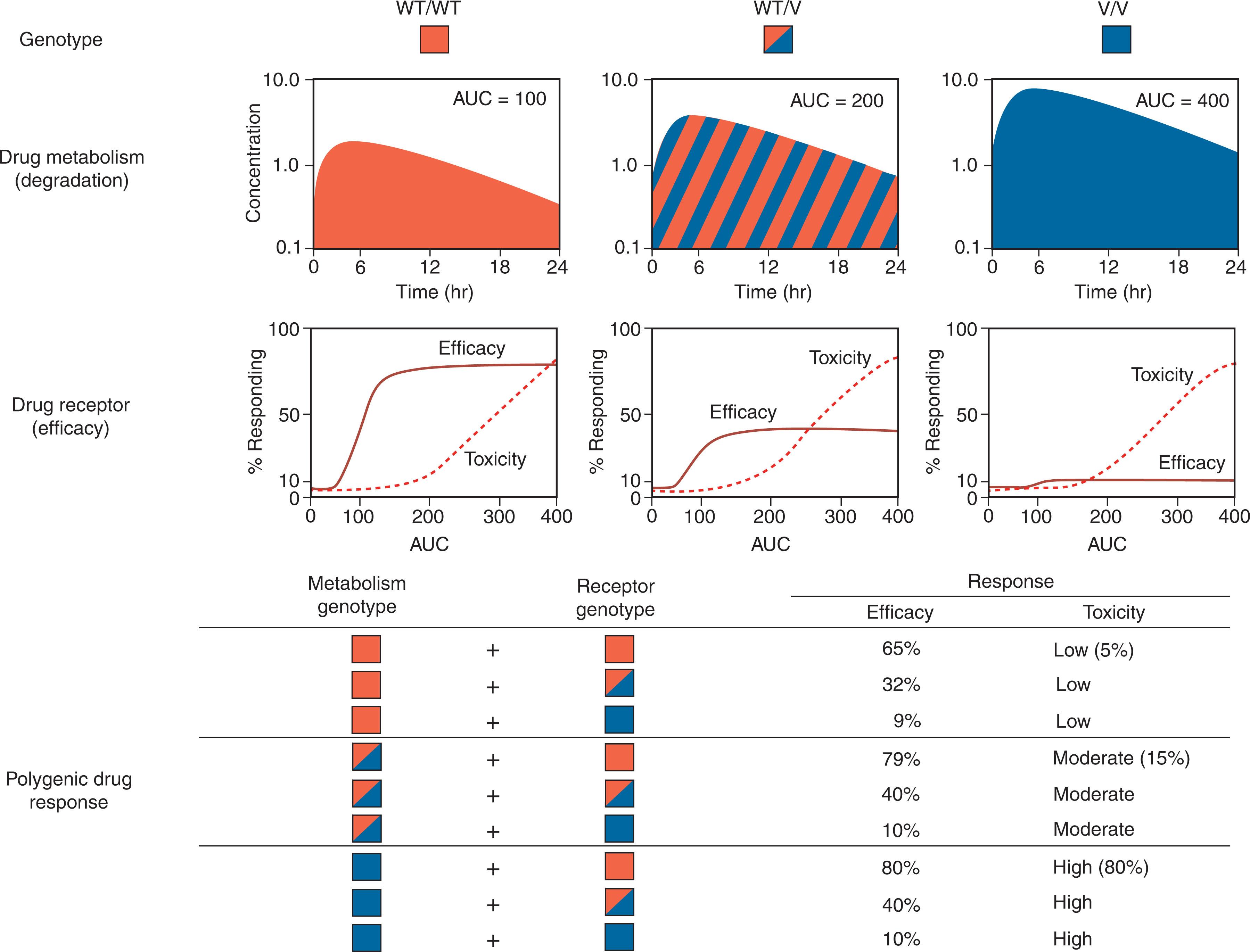
Drug effects are typically determined by the interplay of several gene products that influence the pharmacokinetics and pharmacodynamics of medications. Pharmacokinetics entails characterization of the absorption, distribution, metabolism, and excretion (ADME) of medications. Pharmacodynamics is the relationship between the pharmacokinetic properties of drugs and their pharmacologic effects, either desired or adverse. The ultimate goals of PGx and pharmacoepigenomics in this context are to elucidate the inherited determinants for drug disposition and response to select medications and dosages on the basis of each patient's inherited ability to metabolize, eliminate, and respond to specific drugs. A model of how polygenic variables can determine drug response is illustrated in Fig. 8.1 .
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here