Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
DNA is double-stranded with A-T and C-G pairs of deoxyribonucleotides; it includes exons, introns, and intergenic regions; all nucleated normal cells have the full DNA complement; and DNA is relatively stable. RNA is usually single stranded using uracil in place of thymine; ubiquitous ribonucleases make RNA unstable outside of living cells requiring rapid/immediate sample processing (warm ischemia during surgical resection should especially be avoided); each cell expresses RNAs from only a subset of genes, with dynamic temporal changes, and variable (often short) half-life; and mature RNA lacks introns. DNA is the preferred target for mutation testing and for chimerism analysis after transplant; RNA is the preferred target for detecting gene fusions and performing gene expression analysis. Translocations that do not result in gene fusions will not be detected by RNA analysis, although they may be suspected by aberrant expression of RNA.
Karyotyping requires growing tumor cells in culture. It will detect most rearrangements that result in visible chromosomal alterations ( Figures 23.1A , 23.2A , and 23.3A ).; however, the resolution is limited and “interstitial” rearrangements involving only short segments of chromosomes may be missed. Karyotyping will not identify the involved genes, and it requires viable tumor cells.
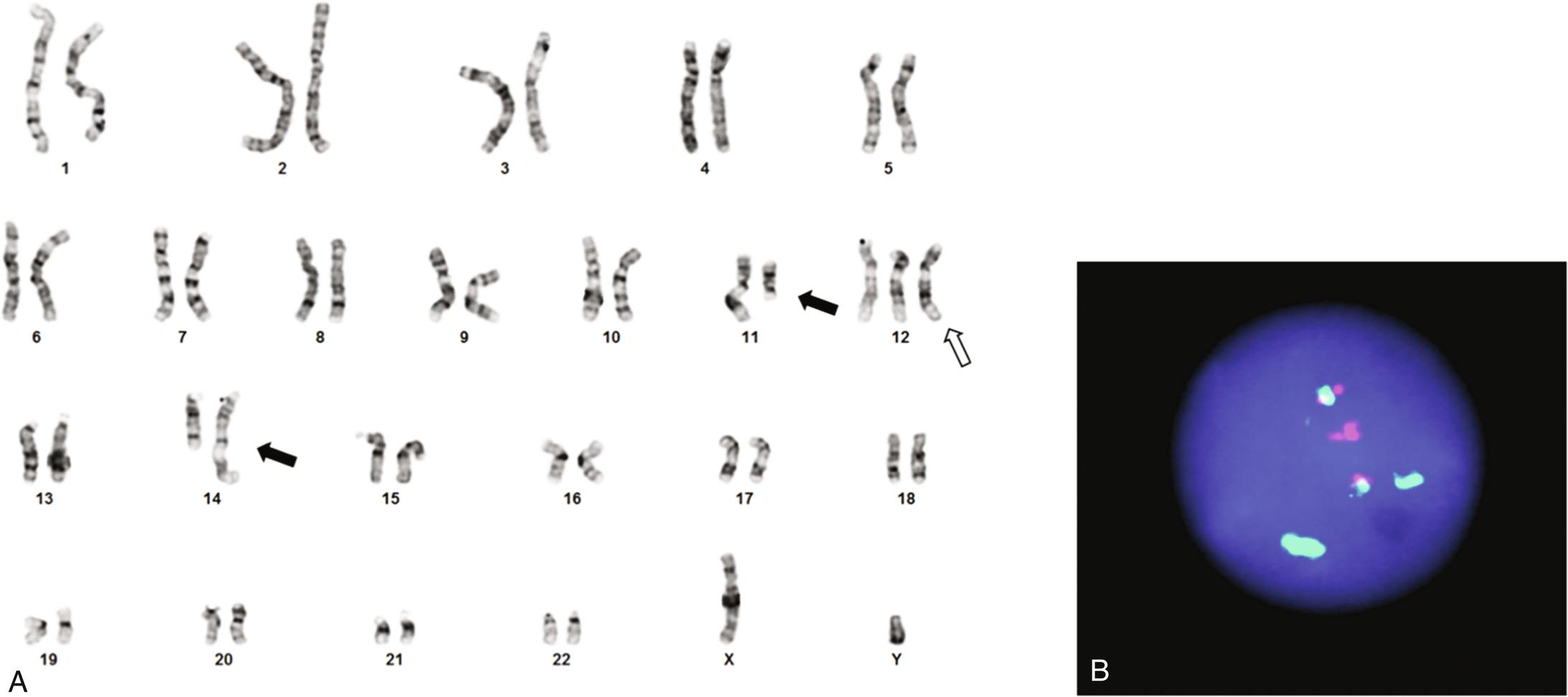
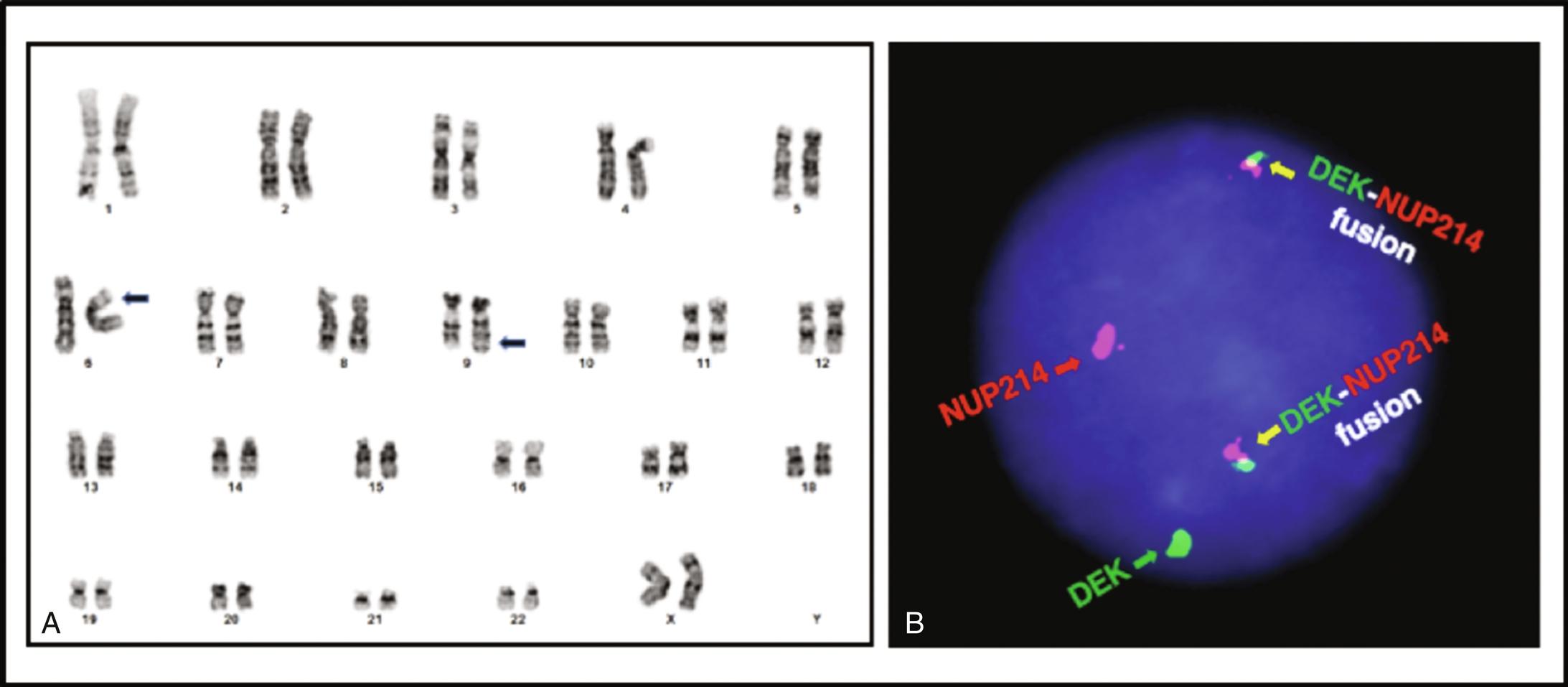
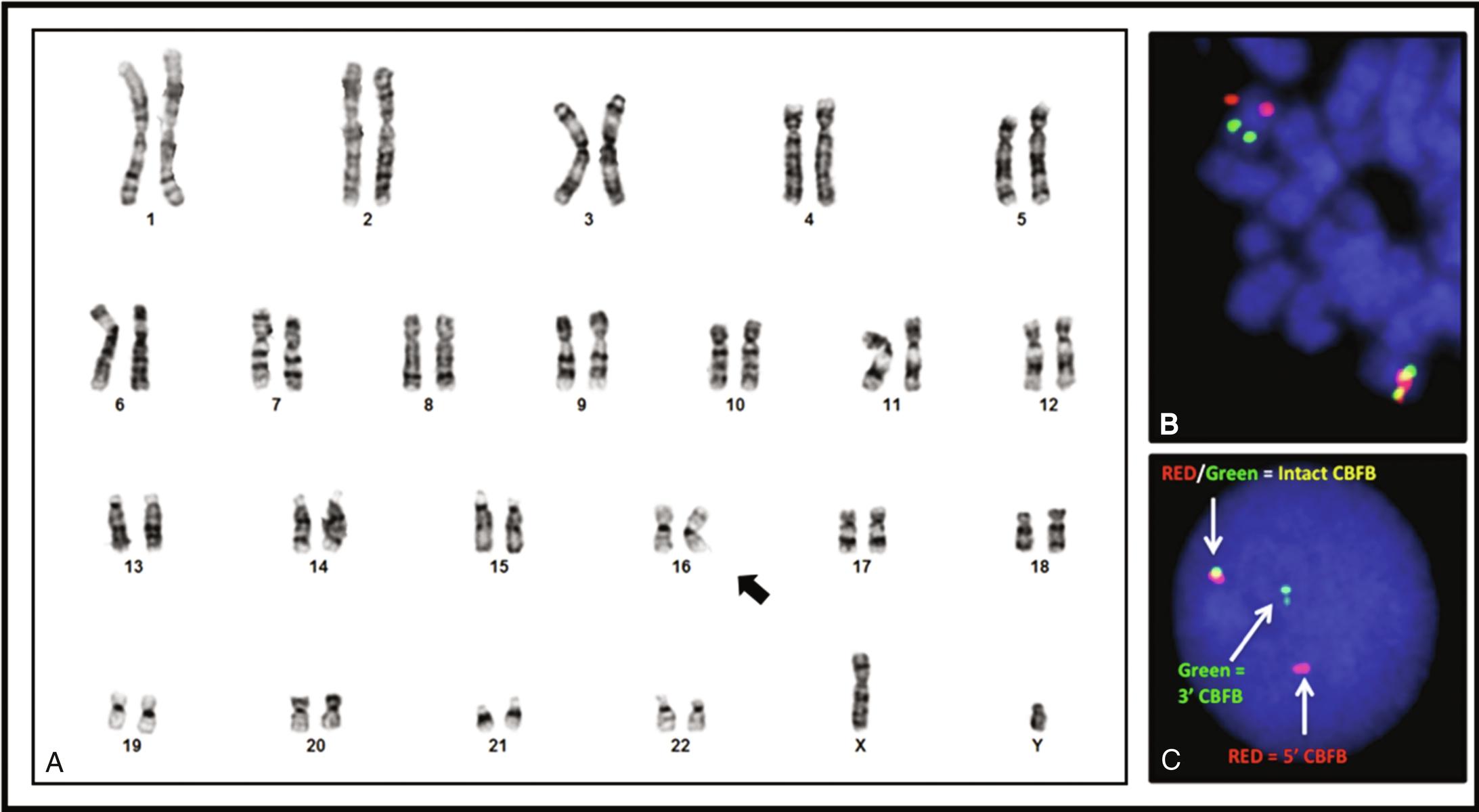
In-situ hybridization uses probes with different colors (fluorescent or chromogenic) targeting distant genomic regions brought together by the rearrangement (“fusion probes”; see Figures 23.1B and 23.2B ) or probes flanking one of the breakpoints, with colors separating as a result of the translocation (“break-apart probes”; see Figure 23.3B–C). A different probe set is needed for each rearrangement, although break-apart probes will detect all rearrangements of a gene that has multiple partners (e.g., EWSR1) without identifying the fusion partner.
Reverse-transcriptase polymerase chain reaction (PCR) can identify rearrangements that create chimeric RNA (e.g., BCR-ABL1 , the various EWSR1 transcripts), but a different primer set and/or test may be needed for each fusion variant. It requires intact RNA and special handling of samples to preserve RNA. Quantitative and highly sensitive tests (tests with low detection limits) can be designed to monitor response to treatment and minimal residual disease.
DNA PCR can detect rearrangement with or without RNA fusions, but DNA breakpoints of rearrangements can vary tremendously from case to case, so only a subset of rearrangements will be detected with each assay. Even multiplexed assays will miss some rearrangements, resulting in variable sensitivity for the various rearrangements.
Targeted next-generation cDNA sequencing using anchored multiplexed PCR can detect RNA fusions of multiple targets at a time and will detect both known and novel partners of targeted genes. It works with partially degraded RNA and with small amounts of RNA. It will only detect fusions involving targeted genes, although modifications allow detection of novel partners.
Whole transcriptome sequencing has the potential to detect all possible rearrangements that result in RNA fusions but is dependent on the level of expression of the fusion RNA. In addition, many nonspecific “fusions” are seen, necessitating careful analysis and, sometimes, confirmatory tests to report novel rearrangements.
Whole-genome sequencing can potentially detect any genomic rearrangement but requires a high tumor percentage in the sample, needs infrastructure to analyze massive amounts of data, and may require additional testing to predict the effects of genomic rearrangements.
High risk: Hypodiploid genome (45 or fewer chromosomes); KMT2A (MLL) rearrangements, most commonly t(4;11)(q21;q23) KMT2A-AFF1 (MLL-AF4 ), t(11;19)(q23.3;p13.3) ( KMT2A-MLLT1) and over forty others; t(9;22)(q34;q11.2) BCR-ABL1 (standard risk with ABL1 inhibitor); Ph-like ( IKZF1I deletions); CRLF2 rearrangements (e.g., t(X;14)(p22;q32)); activating kinase mutations; intrachromosomal amplification of chromosome 21 (iAMP21); MEF2D rearrangements.
Intermediate (standard) risk: t(1;19)(p23;q13.3) TCF3-PBX1 (E2A-PBX1) (standard risk with intensive chemotherapy); t(5;14)(p31;q32) IL3-IGH; ZNF384 rearrangements.
Low risk: ETV6-RUNX1 (TEL-AML1)/t(12;21) (p13;q22) (the most common rearrangement in B-ALL); hyperdiploidy (51–65 chromosomes because of whole chromosome gains); DUX4-IGH gene rearrangements.
The genomic breakpoints involved in BCR-ABL1 translocations all involve intron 1 of ABL1, with three different sets of breakpoints in BCR. The BCR major breakpoint cluster region (M-bcr) in introns 13 and 14 results in fusions of exon 13 or 14 with exon 2 or, less commonly, because of alternative splicing, with exon 3 of ABL1 (e13a2, e13a3, e14a2 and e14a3), which encode the “p210” (approximately 210kd) BCR-ABL1 fusion protein, associated with chronic myeloid leukemia (CML). The minor breakpoint cluster region, in intron 1 of BCR produces the e1a2 and e1a3 fusion transcripts, which encode the p190 BCR-ABL1 fusion protein, characteristic of Philadelphia chromosome positive ALL (Ph + ALL). The least common micro breakpoint cluster in intron 19 (μ-bcr) results in e19a2 and e19a3 fusions, which encode the p230 BCR-ABL1 protein seen in patients with a CML variant with increased neutrophils (“chronic neutrophilic leukemia” or “neutrophilic CML”) and, rarely, typical CML.
Molecular tests, especially RT-PCR based tests, should detect the specific fusions and because of occasional overlap (e.g., CML in lymphoid blast phase presenting as Ph + ALL, or rare p190 positive p210-negative CML), negative results for one fusion may need to be followed up with additional testing for the other fusions.
Activation of the RAS / RAF / MEK / ERK (MAP kinase) pathway is a common finding in Langerhans cell histiocytosis (LCH) and in the non-Langerhans cell histiocytoses, Erdheim Chester disease (ECD), juvenile xanthogranuloma (JXG), and, to a lesser extent, in Rosai Dorfaman disease (RDD). BRAF V600E mutations are predominant in LCH and JXG, followed by MAP2K1 and non-V600E BRAF mutations. NRAS/KRAS mutations predominate in RDD, whereas activation of MAPK signaling in JXG is more heterogenous with CSF1R, KIT, ALK, MET, CSF3R, MAPK2K1, KRAS, and NRAS mutations and fusions involving NTRK1, BRAF, RET, and ALK. Targeting with BRAF and MEK inhibitors is associated with transient responses in tumors with activating mutations in the MAP kinase pathway. Kinase fusions may respond to specific inhibitors.
According to the 2021 International Neuroblastoma Risk Group Staging System (INRGSS), MYCN amplification, a diploid genome (46 chromosomes), and segmental chromosomal alterations worsen risk groups as follows. Excluding fully resected, localized tumors with no imaging defined risk factors, all MYCN amplified tumors are classified as high risk. Among MYCN nonamplified stage M (stage IV) tumors in children younger than 18 months, segmental chromosomal alterations (e.g., 11p losses), diploidy, or unfavorable histology alter the risk category from intermediate to high risk. Activating ALK mutations are unrelated to outcome, but germline ALK mutations and loss-of-function PHOX2B mutations are associated with hereditary neuroblastoma syndromes.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here