Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Pediatric rheumatology owes a great debt to meticulous clinical observations. In the 21st century, however, the practice of pediatric rheumatology requires skills beyond obtaining a thorough history and physical examination. In this chapter, we endeavor to build upon the basic concepts of immunology to introduce and contextualize some of the laboratory methods used to investigate, and increasingly to manage, pediatric rheumatic diseases. This chapter will focus on methodologies related to the quantitation and characterization of RNAs, proteins, microbes, and cells relevant to pediatric rheumatology. Some well-established techniques are already used routinely in day-to-day clinical practice. Others represent more recent technical breakthroughs. Increasingly, collaborative efforts (such as the Accelerating Medicines Partnership RA/SLE) are combining complementary techniques to profile rheumatic immunopathology in an unprecedented level of detail. As the lines distinguishing autoimmunity, autoinflammation, immunodeficiency, and lymphoproliferation continually blur, some techniques once esoteric in other fields are becoming increasingly relevant to practicing rheumatologists. Although a detailed review is beyond the scope of this chapter, it is essential to understand the basic premises, uses, and limitations of these tests to practice competently and use new findings in our rapidly evolving field.
Whether taping blood to the wall for an erythrocyte sedimentation rate or analyzing single-cell RNA sequencing data, clinicians, and investigators should apply concepts relevant to every test to those outlined in this chapter. The concepts of validity, reproducibility, dynamic range, sensitivity, specificity, and positive- and negative-predictive values are discussed elsewhere in this text, but we will highlight these concepts when relevant.
The establishment of normal ranges is particularly relevant in new or experimental biomarker and functional studies. Normal ranges established with only healthy controls frequently lead to the erroneous belief that an abnormal result is specific for one disease. One example of this erroneous belief can be seen in the history of acute phase reactants and their association with infection. Likewise, small or preclinical studies are rarely powered to account for the reality that normal ranges can vary by age, sex, and ethnicity.
Ideally, our measurements would indicate the absolute amount of the targeted substance. Many of our clinical tests report values in terms of mg/dL or cells/mL. Many measurements, however, represent a quantitation relative to a fixed point or standard. Even a typical enzyme-linked immunosorbent assay (ELISA), which reports absolute quantities in units like pg/mL, actually generates this quantitation via reference to the standard curve. Some quantities, particularly messenger RNA (mRNA), are difficult to quantitate absolutely and are routinely expressed in relation to “housekeeping” genes (whose expression is assumed to remain stable across different patients and activation states) or to the total pool of mRNA in a sample (see section titled RNA-sequencing). Savvy observers will recognize that relative values can vary when there are changes in both what one aims to measure (usually the numerator) and what one assumes to be stable.
Hypothesis testing has been productive in moving our field forward and remains a bulwark in the scientific paradigm. However, this paradigm developed in a context where investigators were capable of making only one (or a few) measurements at a time. Increasingly, the profusion of massive multiparameter testing has altered the design and conduct of many experiments. Measurements of the genome, transcriptome, proteome, and metabolome are often sufficiently comprehensive to enable unbiased analyses. This approach has entered everyday practice as we increasingly perform clinical whole exome sequencing (WES; see Chapter 2 ) on patients with suspected monogenic diseases.
Well-designed observational studies employing unbiased measurements often provide rich data sets. We can (and should) use the resulting databases to create and refine hypotheses beyond the capacity of the initial investigators. Many such databases exist and are carefully curated, frequently cited, and clinically useful. Because the ability to use these databases is an increasingly critical skill for investigating and managing rheumatic diseases, we have provided a list of such resources available at the time of printing ( Table 3.1 ) and strongly encourage readers to explore them. Likewise, we must remain alert for new databases as they appear and for updates and additions to existing resources as they become available.
| Resource | Abbr | Description | Link |
|---|---|---|---|
| Genomic | |||
| Genome Aggregation Database | gnomAD | Repository of aggregated human whole exome and genome sequencing | https://gnomad.broadinstitute.org |
| Database of Genotypes and Phenotypes | dbGAP | Archive of sequences and phenotypes from NIH-funded studies | https://www.ncbi.nlm.nih.gov/gap/ |
| ClinVar | ClinVar | Aggregated information about genomic variants in human disease. | https://www.ncbi.nlm.nih.gov/clinvar/ |
| Online Mendelian Inheritance in Man | OMIM | Online catalog of human genes and genetic disorders | https://www.omim.org/ |
| InFevers | – | Curated Registry of Autoinflammatory mutations | https://infevers.umai-montpellier.fr/web/index.php |
| GenomeBrowser | Online visualization tool for nearly any genetic or epigenetic dataset | https://genome.ucsc.edu/index.html | |
| Transcriptional | |||
| Gene Expression Omnibus | GEO | Mandatory repository for all microarray, RNA-seq, and epigenetic data supported by NIH funding | www.ncbi.nlm.gov/geo |
| ArrayExpress | – | Similar to GEO, an archive of such data for European studies | www.ebi.ac.uk/arrayexpress |
| Immunological Genome Project | Immgen | Purified immune cell gene expression. Largely for mouse immune cells | www.immgen.org |
| Human Cell Atlas | – | Massive effort to characterize the transcriptomes (and eventually other ‘omes) of all human cell types | www.humancellatlas.org |
| Genotype Tissue Expression Portal | GTEx | A comprehensive public resource to study tissue-specific gene expression and regulation | www.gtexportal.org |
| BioGPS 88 | – | Web-based portal for exploring and visualizing large studies of cell- and tissue-specific gene expression | www.biogps.org |
| Single Cell Portal | – | Repository of single-cell RNA-seq studies | https://singlecell.broadinstitute.org/single_cell |
| Epigenetics/Proteomics/Multiple | |||
| Encyclopedia of DNA Elements | ENCODE | Massive repository of epigenetic datasets | https://www.encodeproject.org |
| Immunology Database aand Analysis Portal | ImmPORT | Open access platform for NIAID-funded research data sharing | https://www.immport.org/home |
| All RNA and ChiP Sequencing | ARCHS 4 | Archive of GEO NGS data after standardized processing | https://amp.pharm.mssm.edu/archs4/index.html |
| Accelerating Medicines Partnership, RA and SLE | AMP-RA & AMP-SLE | Bulk and scRNA-Seq and CyTOF Data | https://www.niams.nih.gov/grants-funding/funded-research/accelerating-medicines , data viewable at https://immunogenomics.io/ |
| ProteinAtlas | – | Immunohistochemistory and immunofluorescence of mouse and human tissues and cell lines. Includes antibody information | www.proteinatlas.org |
| Universal Protein Resource | UniProt | Suite of databases and tools for assessing protein structure, mutation effects, sequence alignments, BLAST, TrEMBL, … | https://www.uniprot.org/ |
| Tools | |||
| Broad Institute Data, Software, and tools | – | Repository of many tools including Integrative Genomics Viewer (IGV), collections of gene sets (e.g. MSigDB, REACTOME), Gene Set Enrichment Analysis (GSEA), Genome Analysis Toolkit (GATK), and Morpheus | https://www.broadinstitute.org/data-software-and-tools |
| Bioinformatics for Genomics and Proteomics | BioinfoGP | Repository of several data visualization tools | https://bioinfogp.cnb.csic.es/tools |
Technologic advances in how we observe and sequence nucleic acids have enabled an ongoing revolution in genetics and epigenetics. The investigation of genetic and epigenetic causes of pediatric rheumatic diseases is discussed in Chapter 2 , including technologies such as genome-wide association studies (GWAS), WES and whole genome sequencing (WGS), and chromatin immunoprecipitation (ChIP). Understanding these techniques, and the next-generation sequencing (NGS) technology that enables them, will be important for understanding many of the techniques described in this chapter.
Practitioners have been analyzing the relative abundance of different immune cell subsets for decades. We now recognize the existence and interdependent function and regulation of (at least) dozens of different immune cell types.
The separation of cells by their relative density (e.g., Ficoll-based or elutriation) does not yield specific cell populations. However, decades of intensive study have provided both a catalog of antigens to differentiate cell types (e.g., CD19 on B cells vs. CD4 on helper T cells) and monoclonal antibodies of great specificity and affinity for those antigens. Conjugating such antibodies to magnetic beads or other substrates, followed by separation (magnetic or density) and washing, enables the enrichment for certain cell types often to greater than 90% purity. Positive selection refers to enrichment for the cells attached to the beads and generally leads to higher purity, but it can activate the cells. Negative selectio n refers to bead-based removal of undesired cells, leaving the desired cells untouched but generally of lower purity than positive selection. Intact, live, and relatively pure cell populations can then be interrogated for RNA or protein abundance, as described later in this chapter, or subjected to functional testing.
Flow cytometry refers to the process of using a fluid stream to align cells in a single file for analysis. It provides multiparameter, quantitative, single-cell data at increasing economies of scale, time, and cost. A biological sample is incubated with a cocktail of antibodies or other proteins/chemicals that will bind specifically to cellular targets of interest. Such cellular targets include surface proteins, intracellular cytokines or transcription factors, phosphorylated signaling molecules, DNA, and even certain phospholipids. These antibodies are themselves conjugated to a means of their specific detection. Cells are often analyzed at a rate of hundreds to thousands per second, enabling detailed analysis on many large, distinct cell populations within each sample. Different preparation and staining methods enable users to detect and differentiate antigens on a cell’s surface and intracellular antigens (like cytokines and transcription factors). Detection of intracellular antigens typically involves fixation and killing the cells, and thus limits dynamic assessments. Flow cytometry has also been used to analyze cell fragments, platelets, extracellular vesicles (including exosomes), and different microbes. Though tagging antibodies with fluorophores is widely used, antibodies can also be conjugated to heavy metals and detected by mass spectrometry, or to DNA barcodes and detected by NGS, as discussed later in this chapter.
By far the most common method of rendering antibodies detectable by flow cytometry is by labeling an antibody to a fluorophore. A fluorophore is a fluorescent chemical compound that becomes excited at a specific wavelength spectrum, and then reemits light of a higher wavelength spectrum. Flow cytometry was developed in the 1960s and its use has undergone continuous expansion and refinement. In addition to antibody-fluorophore conjugates, cellular components can be made to fluorescence by means other than antibodies including fluorescent chemicals that intercalate with DNA (e.g., 4′,6-diamidino-2-phenylindole [DAPI]), fluorescently labeled proteins known to bind target molecules (e.g., annexin V binding to phosphatidylserine), or cells engineered to express fluorescent proteins (e.g., green fluorescent protein [GFP]). After preparing a single-cell suspension of stained cells, complex fluidics arrange the cells single file within a fluid stream, and this stream passes through a series of lasers ( Fig. 3.1A ). The fluorophore-conjugated antibodies attached to each cell will emit light of a specific spectrum. After excitation by each laser, detectors evaluate the amount of light emitted at specific wavelengths to give an assessment of the abundance of specific fluorophores (and thereby their attached antibodies). This is reported as a relative fluorescence intensity. The brightness of any given target is determined by (1) the abundance of the target on the cell, (2) the affinity of the antibody, and (3) the brightness of the attached fluorophore. Skilled flow cytometrists can manipulate all of these parameters to optimize the reagents used for analysis. The number of different antibodies able to be used simultaneously has expanded with the development of new laser, fluorophore, and detector combinations. Flow cytometry also provides a relative estimation of a cell’s size (forward scatter) and heterogeneity (side scatter) based on its intrinsic reflection of light.
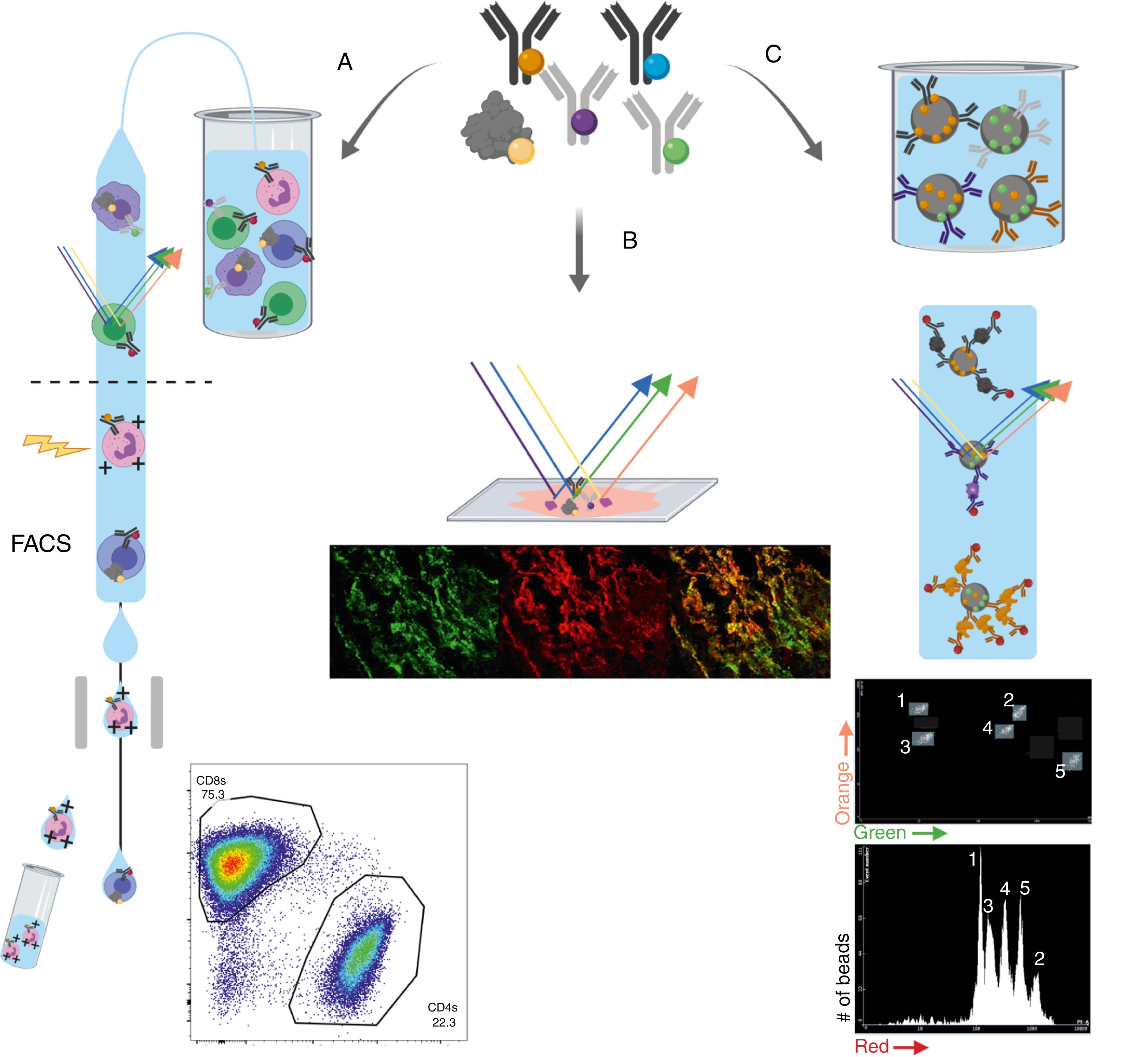
One major advantage of using emitted light to tag cellular targets is the ability to sort cells to an extremely high degree of purity with fluorescence-activated cell sorting (FACS) (see Fig. 3.1A ). Investigators choose which combinations of light patterns best differentiate a population of interest. Cells pass through the laser/detector apparatus and are sorted into droplets. When a cell is found to fulfill target parameters, the droplet into which it was sorted is given a slight electric charge and directed into a separate receptacle when it passes through a capacitor. Live cells can thus be sorted to very high purity based on a combination of dozens of quantitative parameters at a rate of thousands of cells per second.
A great challenge inherent to FACS is compensation. The light emitted by any fluorophore is not restricted to a single wavelength. As one increases the number of fluorophores, the overlap in their light emission spectra is bound to increase. With complex computational methods of resolving this overlap, some current analyzers can simultaneously and quantitatively resolve more than 30 different fluorophores associated with the same cell.
To overcome the limitations of overlapping light emission, investigators recently realized they could conjugate antibodies to heavy metals and analyze the abundance of these heavy metals associated with single cells by mass spectrometry. , Cytometry by time-of-flight (CyTOF), also known as mass cytometry , aligns cells single file, incinerates them, and detects the abundance of various heavy metals on a per-cell basis. Again, this occurs at a rate of hundreds to thousands of cells per second. Because there is effectively no overlap in the time-of-flight (TOF) spectrum of each heavy metal, panels of up to 50 antibodies (and theoretically more) can be used with no need for compensation. This enables an unprecedented level of single-cell quantitative detail. The vast amount of data generated by CyTOF has prompted the development of an expanding array of computational analysis and data visualization tools. CyTOF is currently more costly than flow cytometry due to the challenge of conjugating heavy metals to antibodies and the great number of such antibodies used. Low efficiency is also a challenge, as only a fraction (typically less than half) of the cells that are incinerated are able to be analyzed. Cells analyzed by CyTOF are incinerated and thus unable to be sorted or purified.
Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) is a method of obtaining both single-cell RNA sequencing (scRNA-seq) and single-cell protein abundance data simultaneously. Cells are incubated with antibodies that have been conjugated to DNA bar codes (instead of to fluorophores or heavy metals). Complex fluidics separate and lyse single cells, and NGS captures both the RNA transcriptome and the abundance of DNA bar codes.
Flow cytometry has been used in peripheral blood and synovial fluid in juvenile idiopathic arthritis (JIA), systemic lupus erythematosus (SLE), and countless other diseases to better understand pathogenic mechanisms, differentiate subtypes, and identify biomarkers and therapeutic targets. FACS has enabled flow cytometric findings that distinguish cell types or activation states to be translated into purified populations that can be used for functional assays or assessment of RNA-transcriptomes.
Flow cytometry is used in the routine clinical practice of pediatric rheumatology, including evaluation of lymphocyte subsets as part of an immunodeficiency workup ( Chapter 43 ), assessment of B-cell depletion following treatment with rituximab ( Chapter 14 ), or perforin protein expression in suspected hemophagocytic lymphohistiocytosis ( Chapter 42 ). Early studies in oligo- and polyarticular JIA patients identified more activated T cells in the peripheral blood and notably fewer NK cells. The studies also identified dramatically more innate-like activated B cells (CD5+) and Th1-activated T cells in JIA synovial fluid than in peripheral blood. Later, investigators identified potential markers of extension in the synovial fluid of oligoarticular JIA patients, including CCL5 . In systemic JIA, flow cytometry has been integral in identifying numerical and functional defects in NK cells. More recently, neutrophils from systemic JIA patients, even those whose disease has been quiescent, showed substantial activation manifested as lower expression of the integrin CD62L. Other examples of its investigational utility appear throughout this text in disease-specific chapters.
Quantitating the location and abundance of proteins in biological samples has been a mainstay of investigations in the biological sciences. As with quantitation of RNA, quantitation of one or a few proteins has been performed for decades, and the techniques by which this is accomplished are in routine clinical use. Nearly all of these techniques rely on the strong specificity and affinity of antibodies and are therefore subject to some of the same limitations: No antibody is 100% specific; antibody binding is affected by pH and other environmental variables; and antibodies recognize three-dimensional epitopes that may be affected by changes in the composition of target protein complexes, abnormalities of target protein folding, or steric hindrance of target epitopes.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here