Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The first genome to be sequenced came at a cost of $100 million. Only 13 years later, the price for sequencing an entire genome is nearing $1000 ( Fig. 41-1 ). Rapid increases in the speed and complexity of genomic sequencing technologies, coupled with dramatically decreasing costs, have created an overwhelming array of platforms, methods, and informatics analysis algorithms. In many publications the authors assume the reader is facile with the platform used in the study described, yet most clinicians and researchers have a limited or narrow knowledge of the wide range of methods capable of describing the genomic or proteomic content of a biologic sample. This chapter focuses on describing the basic framework with which to understand most of the common platforms in use today.
Genomic platforms can be classified in many ways, and one can consider the starting material (e.g., deoxyribonucleic acid [DNA], ribonucleic acid [RNA], proteins, and small molecules) or the questions being asked (e.g., copy number, mutations, loss of heterozygosity [LOH], transcript expression, and methylation). The classification scheme shown in Table 41-1 classifies each platform according to the underlying structure, and the remainder of this chapter is organized in this fashion.
| Underlying Structure | “-omics” | Alterations in Disease | Technologies |
|---|---|---|---|
| DNA | Genome | Point mutations | Capillary (Sanger) sequencing Pyrosequencing Genotyping Targeted-sequencing/WES RNA-seq |
| Copy number gains or losses | FISH Array CGH SNP array Targeted-sequencing/WES WGS |
||
| Rearrangements/fusion genes | Karyotyping FISH WGS RNA-seq |
||
| Pathogenic sequences | PCR Microbial arrays WGS RNA-seq |
||
| Epigenome | DNA methylation Histone modifications |
Bisulfite sequencing Methyl-specific PCR ChIP-seq |
|
| mRNA | Transcriptome | Altered transcript expression Altered allele-specific expression Differential alternative splicing |
Microarrays RNA-seq |
| microRNA | Epigenome | Altered transcriptional control | Microarrays |
| Proteins | Proteome | Mutated or deleted proteins Altered posttranslational modification Increased or decreased regulation |
Microarrays Mass spectrometry |
| Small molecules | Metabolome | Modulations in small molecules | Mass spectrometry NMR spectroscopy |
Most of the time the goal of studying DNA is to learn about the genes encoded, including mutations, rearrangements, and changes in copy number. Low-resolution methods prevailed for years as the standard way of understanding copy number variations and rearrangements (e.g., karyotyping and fluorescent in situ hybridization [FISH]), and low-throughput techniques were required to evaluate point mutations and pathogenic sequences (e.g., capillary sequencing, pyrosequencing, polymerase chain reaction [PCR], and microbial arrays). The emergence of high-throughput, high-resolution techniques has revolutionized the study of the genome.
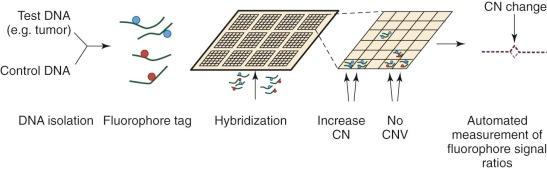
Until the development of comparative genomic hybridization (CGH), chromosomal or subchromosomal gains and losses were discovered though Giemsa banding (“karyotyping”) or FISH. Although similar in resolution to these techniques (5 to 10 megabases ), CGH can be used in an unbiased and agnostic way to detect unbalanced chromosomal abnormalities by competitive FISH using different fluorophores for two different isolates of DNA (e.g., test and control). The technique was also novel because it did not require cells to be undergoing active cellular division. With the popularization of DNA microarrays, a higher resolution technique was developed: array CGH (aCGH). As is common to all microarray techniques, probes (oligonucleotides) are deposited onto a solid support (glass slide), with the resolution dependent on the size of the probes and the genomic distance separating them. DNA from a test sample and control are extracted, labeled with different fluorescent dyes, and applied to the probes ( Fig. 41-2 ). Complementary strands will bind and can be visualized with use of a digital imaging system to quantify the relative amounts of each target bound. The ratio of test to control for each DNA region can then be used to determine copy number variation throughout the genome. Compared with prior methods, aCGH can detect copy number changes at any locus, as long as it is represented in the array. The technique has been productively applied to the study of thousands of diseases and complex traits, and despite the emergence of newer high-resolution methods, aCGH continues to be a significant platform for the study of copy numbers.
Genotyping is simply determining the genetic variation in an individual. Many methods may be used to perform genotyping with varying degrees of throughput. Restriction fragment length polymorphism analysis was productively applied to describe human leukocyte antigen polymorphisms, followed by PCR-based methods using membrane- or bead-bound sequence-specific oligonucleotide probes and sequence-specific priming (reviewed in references and ). For the specific application of identifying a point mutation or allelic variant, genotyping via PCR followed by restriction digestion is a simple approach that has been applied to many diseases and conditions, including fatty acid binding protein mutations in insulin resistance and identification of polymorphisms in thiopurine methyltransferase in leukemia and methylenetetrahydrofolate reductase in childhood acute leukemia.
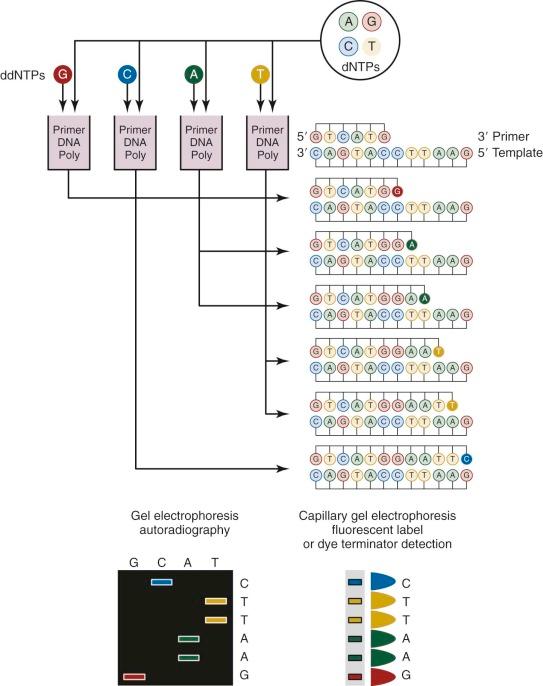
Sanger sequencing, which was first developed in 1977, has been routinely used for more than 25 years for genomic studies. In the classic method ( Fig. 41-3 ), chain-terminating nucleotides are incorporated by DNA polymerase during DNA replication with a synthesized primer specific to the region of interest. Compared with normal deoxynucleoside triphosphates, terminating dideoxynucleoside triphosphates lack a 3′-hydroxyl group, and DNA polymerase is unable to create the phosphodiester bond between two nucleotides, thus halting transcription. Four DNA replication samples are prepared, each with a different radiolabeled or fluorescently tagged dideoxynucleoside triphosphate, and the resulting DNA products are separated by electrophoresis and visualized by autoradiography or ultraviolet light. Current instruments use dye terminators instead of fluorescent labels, resulting in faster and more accurate readings. The relatively high cost for reagents and the nonautomated nature of traditional Sanger sequencing have been largely obviated by the development of microfluidic technology. All of the steps for Sanger sequencing are carried out on a small chip using nanoliter volumes. This “lab-on-a-chip” or “sequencing-on-a-chip” technique increases speed and accuracy while decreasing costs. Despite limited resolving power for the first 25 to 50 bases and read lengths fewer than 1000 bases, Sanger sequencing is still being used for smaller scale projects or when a long contiguous read is desired.
In pyrosequencing, liberated pyrophosphates from nucleotide incorporation are detected and the DNA sequence is determined by light emitted upon nucleotide incorporation. Because only one nucleotide at a time is presented to the DNA template and DNA polymerase, the base responsible for the emitted light is the one incorporated into the growing strand. The pyrophosphate release during the reaction is converted to adenosine triphosphate, fueling a luciferase-based fluorescent reaction. Although the reads are shorter (300 to 500 base pair [bp]), multiple reactions can be detected simultaneously, thereby increasing throughput.
A microarray is simply a small piece of DNA (the “probe”) that is attached to a solid support, which is usually a glass, plastic, or silicon chip (e.g., an Affymetrix [Santa Clara, CA] genome chip, gene array, or DNA chip) or polystyrene beads (Illumina, San Diego, CA). The process of “printing” the array refers to the deposition of the DNA onto the solid support, either through “spotting” (complementary DNA [cDNA] microarray ) or by synthesizing the oligonucleotides directly onto the array surface. For a spotted array, the probes are produced beforehand, either through the production of a cDNA library, by PCR, or by the generation of oligonucleotides that are then “spotted” directly onto the support surface, often by a robot.
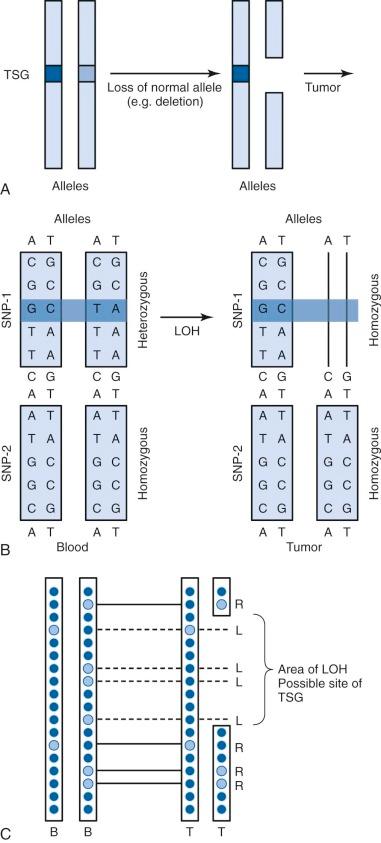
A single nucleotide polymorphism (SNP) is any sequence variation in the genome for which both alleles occur at a relatively high frequency. The vast majority do not appear to confer functional consequences. Although they are located most often in noncoding regions, SNPs can occur anywhere throughout the genome. More than 50 million SNPs have been cataloged in the SNP database. High-density SNP arrays can be used for genetic linkage studies to map disease loci and complex traits. Because SNP arrays can detect slight differences between individual genomes, the polymorphisms detected can be used to characterize disease susceptibility or drug effectiveness. A comparison of intensities of DNA-bound SNP probes can be used to determine the relative DNA copy number for a given locus based on the SNP map. A specific application of SNP arrays is in detection of LOH in tumors and other malignancies ( Fig. 41-4 ). When examining paired blood and tumor samples, SNPs detected as heterozygous in blood and homozygous in tumor (LOH) may be part of a region where a normal copy of a tumor suppressor gene was lost. A special case of LOH called uniparental disomy is a copy-neutral gene conversion. Instead of a deletion leading to the loss of the normal allele, a nondisjunction event results in LOH without a change in copy number. Although undetectable by FISH or karyotyping, this important type of LOH can be inferred from an SNP virtual karyotype.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here