Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The immune system, the function of which is to protect against infections, is composed of two branches: a phylogenetically earlier one called innate immunity and a more recently evolved one called adaptive immunity. Innate and adaptive immunity are not two separate compartments but an integrated system of host defense, sharing bidirectional interactions fundamental to both the inductive phase and the effector phase of the immune response. The innate immune system constitutes the first line of host defense in response to infection and therefore plays a crucial role in the early recognition and triggering of the proinflammatory response to invading pathogens. The adaptive immune system, on the other hand, is responsible for elimination of pathogens in subsequent phases of infection, the maintenance of immunological tolerance, and the generation of immunological memory.
The cells of the immune system originate from pluripotent hematopoietic stem cells that give rise to stem cells of more limited potential (lymphoid and myeloid precursors). These more limited stem cells then give rise to a wide array of cell types that comprise the immune system. This complex system functions by means of an intricate network of cellular interactions involving cell surface proteins (both ligands and receptors) and soluble mediators such as cytokines.
The innate immune system is the first line of defense against microorganisms and is conserved in plants and animals. It is phylogenetically ancient compared with adaptive immunity, which exists only in vertebrates. The principal components of innate immunity are (1) physical and chemical barriers such as epithelia and antimicrobial substances produced at epithelial surfaces; (2) circulating effector proteins such as the complement components and cytokines; (3) cells with innate phagocytic activity such as neutrophils, macrophages, and dendritic cells; and (4) cells that recognize “altered self” such as natural killer (NK) cells.
Innate immune proteins and cell surface receptors recognize highly conserved structures characteristic of microbial pathogens that are not present in mammalian cells. The binding of microbial structures to these receptors triggers phagocytes to engulf the pathogen and induces cytokines, chemokines, and costimulators that recruit and activate antigen-specific lymphocytes and initiate adaptive immune responses. Thus innate immunity not only represents an effective early defense mechanism against infection but also provides the “warning” of the presence of an infection against which a subsequent adaptive immune response has to be mounted.
The primary cells of the phagocyte system originate from a common lineage in the bone marrow, circulate in the blood in inactive form, and are recruited and activated in the peripheral tissues in response to infection, tissue injury, or other proinflammatory stimuli. Monocytes are the classical example of immature circulating phagocytes, characterized by a granular cytoplasm with many phagocytic vacuoles and lysosomes. Once they enter in the tissues, monocytes can mature into macrophages. Macrophages are additionally seeded into tissues from the yolk sac during embryogenesis and replenished from local yolk sac–derived stem cells. Macrophages are strategically placed in all organs and tissues where they act as “sentinels” together with dendritic cells. In fact, one of their major roles is to recognize and respond to microbes and to amplify the response against a potentially harmful stimulus. Depending on the tissues in which they are found, macrophages are known by a number of different names (e.g., Kupffer cells in the liver, microglial cells in the central nervous system, and alveolar macrophages in the airways). These cells are the prototypical effector cells of innate immunity. Once activated, macrophages initiate a number of crucial events, which include phagocytosis and destruction of ingested microbes and production of proinflammatory cytokines and other mediators of inflammation that lead to further recruitment of cells of innate immunity (monocytes, neutrophils) and provide signals to cells (T and B cells) of adaptive immunity.
Dendritic cells (DCs) are another major phagocytic cell type of innate immunity, specialized to present phagocytosed antigen on major histocompatibility complex (MHC) molecules to T cells. DCs arise from bone marrow precursors and can develop into a number of specific lineages, each with unique characteristics in the manner in which they present their antigen. These cells are also capable of copious cytokine production in response to stimulation by pathogens or tissue damage.
Neutrophils are the most abundant type of circulating leukocytes. Their nucleus is segmented into three to five lobules, hence the term polymorphonuclear leukocytes. The cytoplasm is characterized by the presence of two types of granules. So-called specific granules contain enzymes, including lysozyme, elastase, and collagenase. The azurophilic granules are lysosomes containing enzymes and microbicidal substances. Neutrophils are typically the first cells to enter a site of infection and represent the most prevalent cell type in the early phases of an antimicrobial response. Within 1 or 2 days, neutrophils are almost completely replaced by recruited monocytes–macrophages, which represent the dominant effector cells in the later stages of inflammation.
The innate immune response relies on recognition of evolutionarily conserved structures on pathogens, termed pathogen-associated molecular patterns (PAMPs), through a limited number of germ line–encoded pattern recognition receptors (PRRs) ( Table 4.1 ). Among them the family of Toll-like receptors (TLRs) has been studied most extensively. , PAMPs are characterized by being invariant among entire classes of pathogens and distinguishable from “self.” This characteristic allows a limited number of germ line–encoded PRRs to detect the presence of many microbial infections. Finally, because PAMPs are essential for microbial survival, mutations or deletions of PAMPs are lethal, reducing the possibility that microbes undergo PAMP mutations in order to escape recognition by the innate system.
| Molecular pattern | Origin | Receptor | Main Effector Function |
|---|---|---|---|
| LPS | Gram-negative bacteria | TLR4, CD14 | Macrophage activation |
| Unmethylated CpG nucleotides | Bacterial DNA | TLR9 | Macrophage, B-cell, and plasmacytoid cell activation |
| Terminal mannose residues | Microbial glycoprotein and glycolipids | (1) Macrophage mannose receptor (2) Plasma mannose-binding lectin |
Phagocytosis Complement activation opsonization |
| LPS, dsRNA | Bacteria, viruses | Macrophage scavenger receptor | Phagocytosis |
| Zymosan | Fungi | TLR2, Dectin-1 | Macrophage activation |
| dsRNA | Viral | TLR3, RIG-I ∗ | IFN type I production |
| ssRNA | Viral | TLR7/8, MDA5 ∗ | IFN type I production |
| N-formylmethionine residues | Bacteria | Chemokine receptors | Neutrophil and macrophage activation and migration |
| MDP | Gram-positive and gram-negative bacteria | NOD2 ∗ , NALP1 ∗ | Macrophage activation |
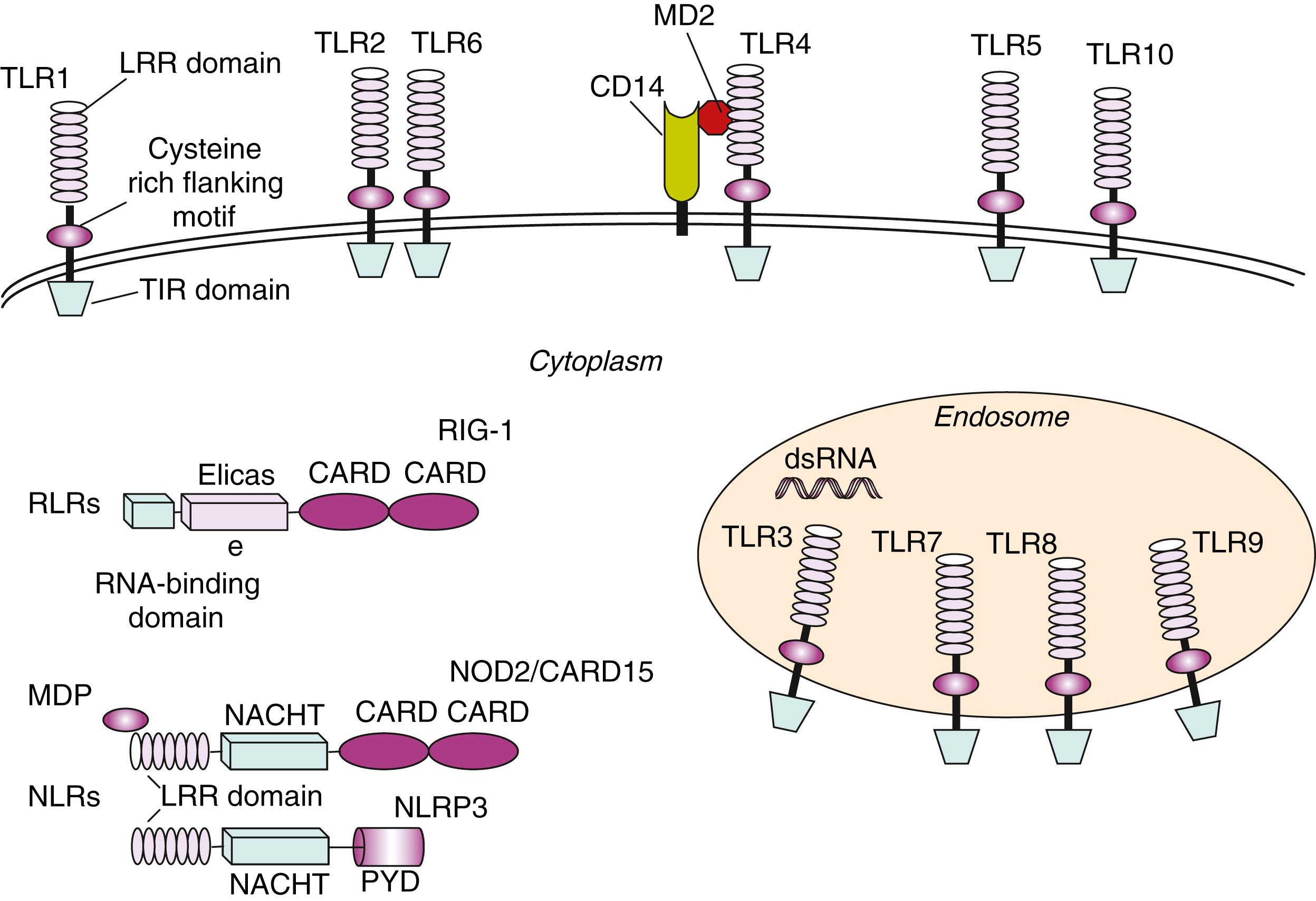
Many classes of PRRs are present on the surface of cells of the innate immune system where they act as “tissue sentinels” through the continuous monitoring of peripheral tissues for the possible invasion of microbial pathogens. TLRs are a large class of PRRs characterized by an extracellular leucine-rich repeat (LRR) domain and an intracellular Toll/IL-1 receptor (TIR) domain ( Fig. 4.1 ). To date, 13 TLRs have been identified in humans, and they each recognize distinct PAMPs derived from various microbial pathogens, including viruses, bacteria, fungi, and protozoa ( Table 4.2 ). ,
| TLR | Cellular localization | Ligand(s) | Microbial Source |
|---|---|---|---|
| TLR1 | Cell surface | Lipopeptides | Bacteria, mycobacteria |
| TLR2 | Cell surface | Zymosan Peptidoglycans Lipoteichoic acids Lipoarabinomannan Porins Envelope glycoproteins |
Fungi Gram-positive bacteria Gram-positive bacteria Mycobacteria Neisseria Viruses (e.g., measles, HSV, CMV) |
| TLR3 | Endolysosomal compartment | dsRNA | Viruses |
| TLR4 | Cell surface and endolysosomal compartment | LPS Lipoprotein HSP60 Fusion protein |
Gram-negative bacteria Many pathogens Chlamydia pneumoniae RSV |
| TLR5 | Cell surface | Flagellin | Bacteria |
| TLR6 | Cell surface | Diacyl lipopeptides Lipoteichoic acid |
Mycoplasma Gram-positive bacteria |
| TLR7 | Endolysosomal compartment | ssRNA and short dsRNA | Viruses and bacteria |
| TLR8 | Endolysosomal compartment | ssRNA and short dsRNA | Viruses and bacteria |
| TLR9 | Endolysosomal compartment | Unmethylated CpG DNA | Bacteria, protozoa, viruses |
| TLR10 | Cell surface | Unknown | – |
| TLR11 | Endolysosomal compartment | Profilin and flagellin | Apicomplexan parasites |
| TLR12 | Endolysosomal compartment | Profilin | Apicomplexan parasites |
| TLR13 | Endolysosomal compartment | Bacterial 23S rRNA | Gram-negative bacteria |
Certain TLRs ( TLR1 , - 2 , - 4 , - 5 , - 6 , and - 10 ) are expressed at the cell surface and mainly recognize bacterial products unique to bacteria, whereas others ( TLR3 , - 7 , - 8 , - 9 , - 11 , - 12 , - 13 ) are located almost exclusively in intracellular compartments, including endosomes and lysosomes ( Fig. 4.1 ) and are specialized in recognition of nucleic acids, with self- versus nonself-discrimination provided in part by the localization of the ligands in an unusual compartment (endosome), as well as unique structures of pathogen versus host nucleic acid (methylation of CpG motifs). , ,
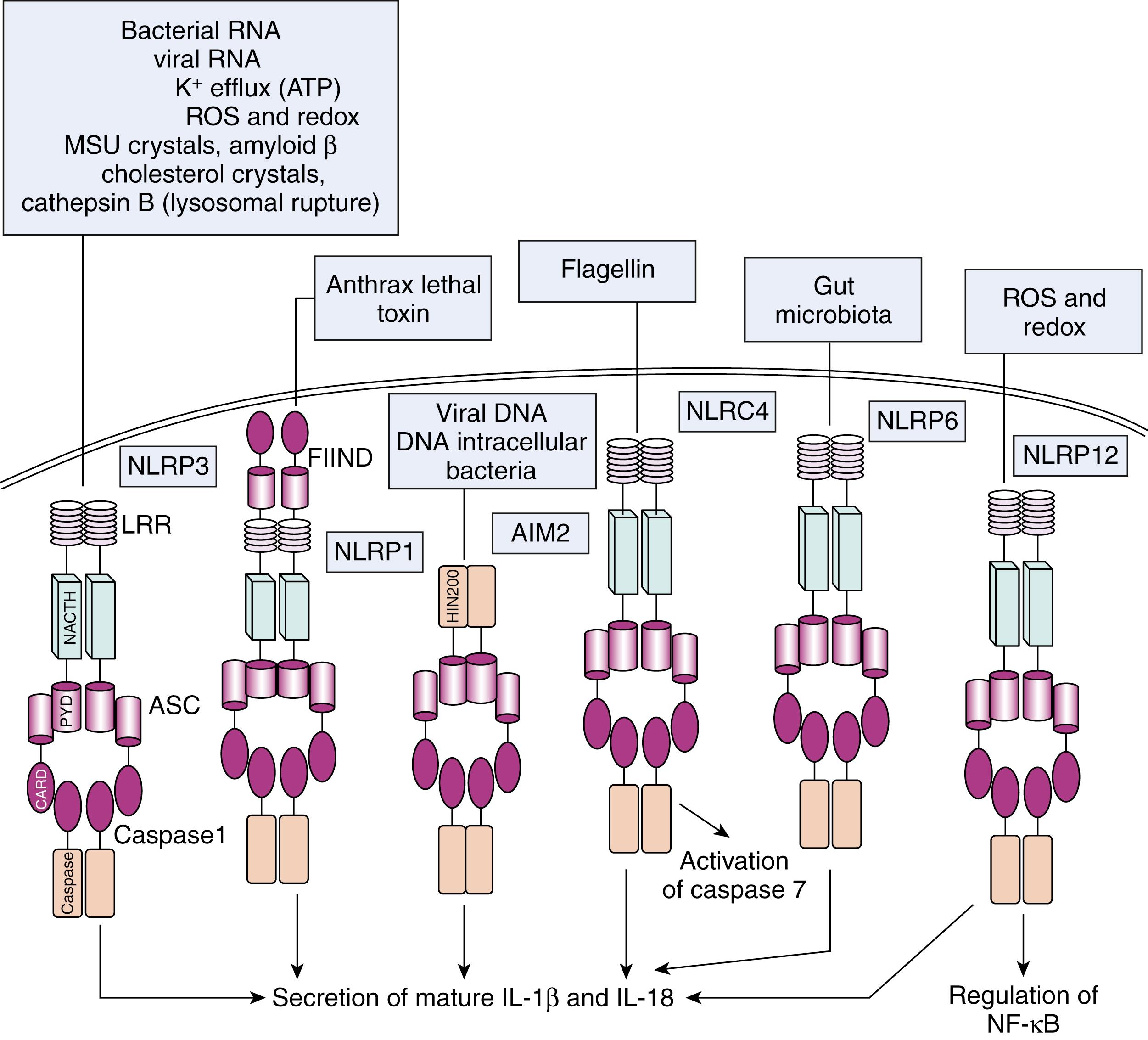
The key cell types expressing TLRs are antigen-presenting cells (APCs), including macrophages, DCs, and B lymphocytes. Ligand binding to TLRs through PAMP–TLR interaction induces an antimicrobial proinflammatory response that is also able to involve and orient the adaptive immune system. In addition to transmembrane receptors on the cell surface and in endosomal compartments, there are intracellular (cytosolic) receptors that function in the pattern recognition of bacterial and viral pathogens. These include nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and the intracellular sensors of viral nucleic acids, such as RIG-I (retinoic-acid-inducible gene I) or melanoma differentiation-associated gene 5 ( MDA5 ), grouped under the term RIG-I-like receptors (RLRs) ( Fig. 4.1 ). NLRs are a family of about 23 intracellular proteins with a common protein-domain organization but diverse functions ( Table 4.3 ). NLRs are composed of a variable N-terminal effector region consisting of a caspase recruitment domain (CARD), pyrin domain (PYD), acidic domain, or baculovirus inhibitor repeats (BIRs), a centrally located NOD (or NACTH) domain that is critical for activation, and C-terminal leucine-rich repeats (LRRs) that sense PAMPs ( Fig. 4.1 ). The NOD and NLRP (NLRs containing a pyrin domain) subfamilies are the most characterized among NLRs.
| Name ∗ | Other Names | Microbial Motifs Recognized | NLR Family |
|---|---|---|---|
| CIITA | NLRA, C2TA | NLRA | |
| NAIP | NLRB1, BIRC1 | NLRB | |
| NOD1 NOD2 |
NLRC1, CARD4, NLRC2, CARD15, BLAU |
GM-tripeptide MDP |
NLRC |
| NLRC3 NLRC4 NLRC5 |
NOD3 CARD12, IPAF NOD27 |
Flagellin from Salmonella, Shigella, Listeria, Pseudomonas | |
| NLRP1 NLRP2 NLRP3 NLRP4 NLRP5 NLRP6 NLRP7 NLRP8 NLRP9 NLRP10 NLRP11 NLRP12 NLRP13 NLRP14 |
NALP1, CARD7 NALP2, PYPAF2 NALP3, CIAS1, Cryopyrin, PYPAF3 NALP4, PYPAF4 NALP5, PYPAF8 NALP6, PYPAF5 NALP7, PYPAF3 NALP8, NOD16 NALP9, NOD6 NALP10, NOD8 NALP11, PYPAF6, NOD17 NALP12, PYPAF2, Monarch1 NALP13, NOD14 NALP14, NOD5 |
MDP Bacterial RNA, viral RNA, uric acid crystals, LPS, MDP |
NLRP |
| NLRXI | NOD9 | NLRX |
∗ According to the Human Genome Organization Gene Nomenclature Committee (HGNC) GM, Tripeptide-GlcNAc-MurNAc tripeptide; LPS, lipopolysaccharide; MDP, muramyl dipeptide.
The proteins of the NOD subfamily, NOD1 and NOD2, are involved in sensing bacterial molecules derived from synthesis and degradation of peptidoglycan. Whereas NOD1 recognizes diaminopimelic acid produced primarily by gram-negative bacteria, NOD2 is activated by muramyl dipeptide (MDP), a component of both gram-positive and gram-negative bacteria. The NLRP subfamily of NLRs has 14 members, and at least some of these are involved in activation of the interleukin (IL)-1 family of cytokines, which includes IL-1β and IL-18. These cytokines are synthesized as inactive precursors that are cleaved by the proinflammatory caspases, such as caspases 1, 4, and 5. These caspases are activated in a multisubunit complex called the inflammasome (see The Inflammasomes and Their Role in the Secretion of IL1β).
Most PRRs sense not only pathogens but also misfolded/glycated proteins or exposed hydrophobic portions of molecules released by injured cells termed damage-associated molecular patterns (DAMPs). , DAMPs, including high-mobility group box 1 protein (HMGB-1), heat-shock proteins (HSPs), uric acid, altered matrix proteins, and S100 proteins, represent important danger signals that are released from activated or necrotic cells and mediate inflammatory responses through TLRs or NLRPs, or other specific receptors like RAGE (receptor for advanced glycation end-products). The term alarmins has also been proposed for DAMPs released during necrotic or inflammatory based cell death. A prototypic DAMP, the nuclear protein HMGB-1, is either passively released by necrotic cells or actively secreted by some activated cells. IL-33, another example of an alarmin, is a member of the IL-1 cytokine family that has been implicated in diseases such as hemophagocytic lymphohistiocytosis.
S100A8, S100A9, and S100A12 (also named calgranulins) are calcium-binding proteins expressed in the cytoplasm of phagocytes and secreted by activated monocytes or neutrophils. Once released from cells, calgranulins exert numerous extracellular functions. Secreted S100A8/A9 complexes (also known as calprotectin) bind specifically to endothelial cells and directly activate the microvascular endothelium, leading to loss of barrier function, apoptosis of endothelial cells, upregulation of thrombogenic factors, and an increase of junctional permeability. S100A8/A9 and S100A12 upregulate expression and affinity of the integrin receptor on neutrophils and facilitate their adhesion to fibrinogen and to fibronectin and the adhesion of monocytes to the endothelium in vitro. In addition, the S100A8/A9 complex and S100A8 itself bind to and signal directly through the lipopolysaccharide (LPS) receptor complex including TLR4 , MD2, and CD14. The binding to these receptors induces the activation of a number of intracellular proinflammatory pathways (see later in the chapter).
The binding of microbes to phagocytes through PRRs initiates the process of phagocytosis of microorganisms and subsequent killing in phagolysosomes. Activation of phagocytes through PRRs also induces effector molecules such as inducible nitric oxide synthase and antimicrobial peptides that can directly destroy microbial pathogens. This is particularly true for polymorphonuclear neutrophils, which are major contributors to the early innate immune response. Their capacity of phagocytosis exceeds that of macrophages, but their capacity to synthesize RNA and proteins is low. Neutrophils are a major source of oxidants, including reactive oxygen species (ROS) and reactive nitrogen (RNS) species, which participate in regulation of the immune response and intracellular killing of bacterial pathogens. The main source of ROS in neutrophils is the membrane-bound enzyme complex nicotinamide adenine dinucleotide phosphate (NADPH) oxidase.
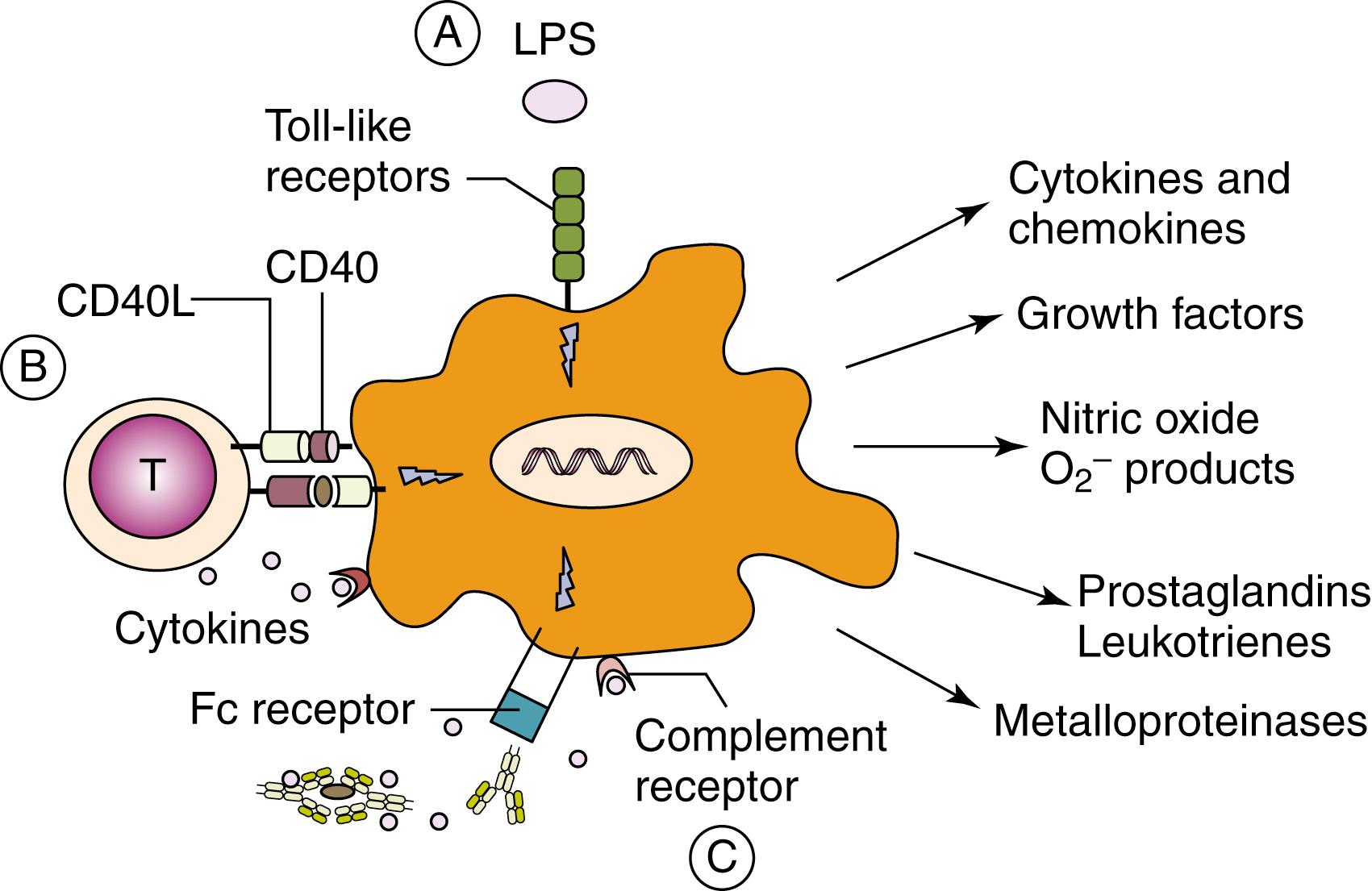
Peptides are generated from microbial proteins and presented by professional APCs, such as DCs to T cells, to initiate adaptive immune responses. After antigen uptake, DCs become activated and migrate to regional lymph nodes to present antigenic peptides in the context of relevant MHC molecules. During this process, phagocytosis, upregulation of costimulatory molecules (including CD80, CD86, and CD40, and antigen-presenting MHC molecules), changes in chemokine receptor expression, and cytokine secretion are all events that are regulated through the recognition of pathogens by PRRs expressed on DCs ( Fig. 4.2 ).
Upon engagement of TLRs by individual PAMPs, a number of different signaling pathways are triggered. Signal transduction is mediated initially by a family of adaptor molecules, which at least in part determines the specificity of the response. , Recruitment of one or several adaptor molecules by a given TLR is followed by activation of downstream signal transduction pathways via phosphorylation, ubiquitination, or protein–protein interactions, ultimately culminating in activation of transcription factors that regulate the expression of genes involved in inflammation and antimicrobial host defense ( Fig. 4.3 ). TLR-induced signaling pathways can be broadly classified on the basis of their utilization of different adaptor molecules, that is, dependent on or independent of the myeloid differentiation primary response 88 (MyD88) adaptor protein or the TIR domain-containing adaptor inducing IFN-γ (TRIF) adaptor protein and their respective activation of individual kinases and transcription factors. , Three major downstream signaling pathways responsible for mediating TLR-induced responses include (1) nuclear factor κB (NF-κB), (2) mitogen-activated protein kinases (MAPKs), and (3) IFN regulatory factors (IRFs). Whereas NF-κB and MAPKs play central roles in induction of a proinflammatory response, IRFs are essential for stimulation of IFN production , ( Fig. 4.3 ).
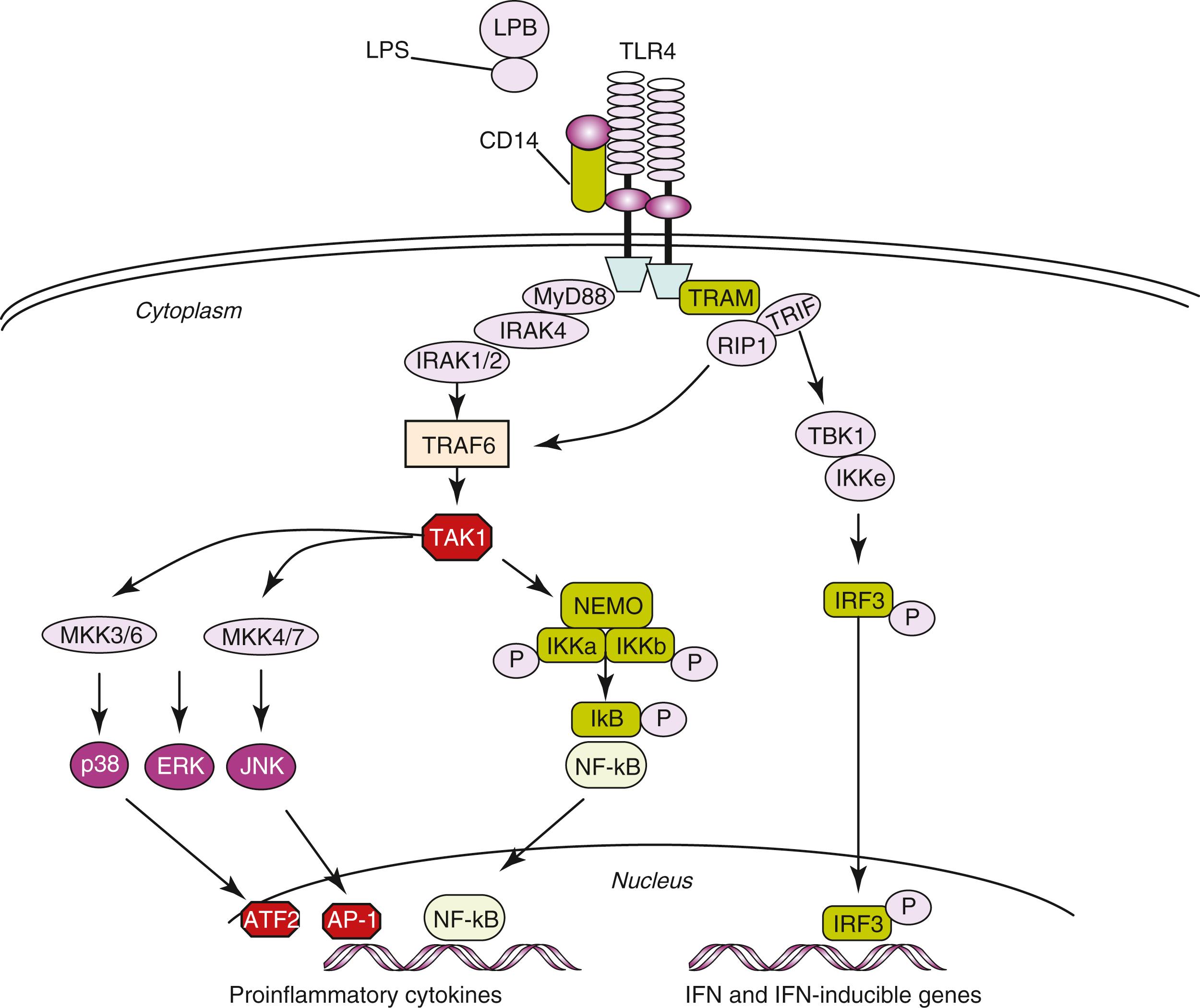
After ligand binding, TLRs dimerize and undergo conformational changes required for the subsequent recruitment of cytosolic TIR domain-containing adaptor molecules. MyD88 is involved in signaling triggered by all TLRs, with the exception of TLR3 , and plays a major role in TLR-induced signal transduction. In response to TLR4 stimulation by an appropriate PAMP, MyD88 associates with the cytoplasmic part of the receptor and recruits members of the IL-1 receptor (IL-1R)-associated kinase (IRAK) family. IRAK1 or IRAK2 associate with TRAF6 , which catalyzes the synthesis of transforming growth factor-activated protein kinase 1 ( TAK1 ) and the IκB kinase (IKK) subunit NF-κB essential modifier (NEMO). TAK1 then stimulates two distinct pathways involving the IKK complex and the MAPK pathway, respectively , (see Fig. 4.3 ).
NF-κB exists in an inactive form in the cytoplasm physically associated with its inhibitory protein inhibitor of NF-κB (IκB). Upon inflammatory stimuli, IκB is phosphorylated and degraded releasing NF-κB dimers, which translocate to the nucleus. Phosphorylation of IκB is performed by IKK. NF-κB binds to a variety of promoters or enhancers of target genes in the nucleus leading to increased transcription and expression. , ,
MAPKs are an important kinase family involved in rapid downstream inflammatory signal transduction resulting in activation of several nuclear proteins and transcription factors. MAPK pathways are activated through sequential phosphorylations, beginning with activation of MAPK kinase kinase (MAPKKK), which phosphorylates and activates MAPK kinase (MAPKK), which in turn activates MAPK. MAPK-activated pathways include p38, JNK, and ERK. p38 and JNK phosphorylate activate transcription factors such as ATF-2 and AP-1, which are necessary for the upregulation of several proinflammatory molecules.
Cytosolic PRRs like NLRs and intracellular sensors of viral nucleic acids (RIG and DAI) exert the same function played by the membrane-bound PRRs. For example, the stimulation of NOD1 or NOD2 by bacterial-derived peptidoglycan fragments results in the activation of NF-κB and MAPKs, which drive the transcription of numerous genes involved in both innate and adaptive immune responses. , ,
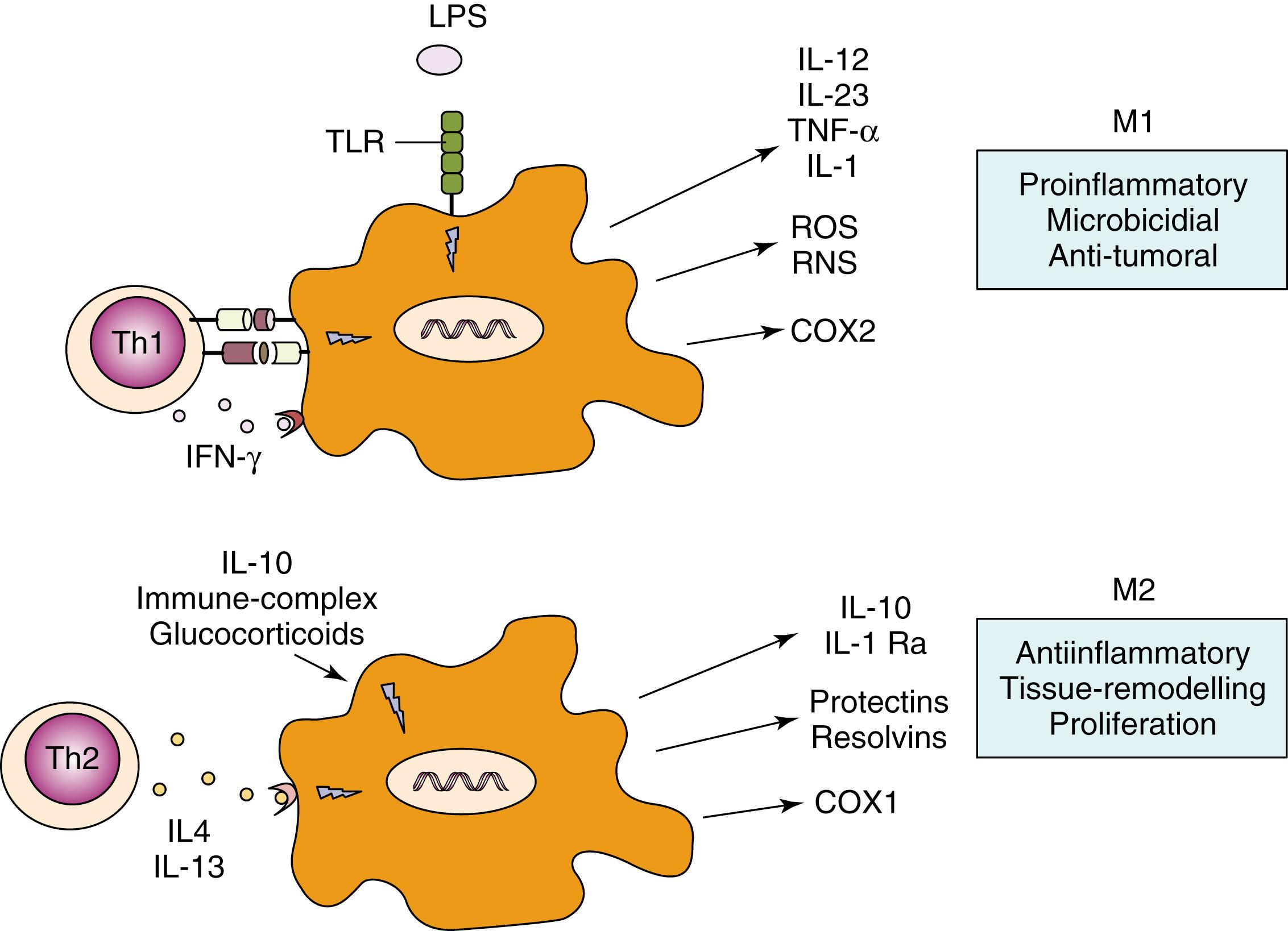
Innate immune cells can also be activated through interaction with a number of other cells and soluble factors of the immune system. Thus macrophage and neutrophils are activated by immune complexes and complement fragments through their binding with immunoglobulin and complement receptors expressed on their cell surfaces ( Fig. 4.2 ). The functional activity of macrophages after stimulation of PRRs is highly influenced by the environmental conditions and by their interaction with other cells. In response to signals derived from microbes, damaged tissues, or activated lymphocytes, it has been suggested that macrophages may develop into one of two different states: the classically activated M1 phenotype or the alternatively activated M2 phenotype ( Fig. 4.4 ). T helper(Th)1-related cytokines like IFN-γ (see Chapter 5 ), as well as microbicidal stimuli, polarize macrophages to an M1 phenotype. M1 macrophages produce high levels of IL-12 and IL-23 and other molecules important in inflammatory, microbicidal, and tumoricidal activities ( Fig. 4.4 ). In contrast, Th2 cytokines like IL-4 and IL-13 (see Chapter 5 ) polarize macrophages to an alternatively activated (or M2) phenotype, characterized by immunomodulatory mediators such as IL-10, IL-1 decoy receptor (IL-1R2), and IL-1 receptor antagonist (IL-1Ra). M2 macrophages dampen inflammation, promote tissue remodeling and repair, help in parasite clearance, and possess immunoregulatory functions, which may contribute to their purported role in suppressing tumor immunosurveillance ( Fig. 4.4 ). These macrophage fates are likely to be very plastic, with a single macrophage changing between M1 and M2 functions in response to changes in the local environment.
There are a number of intracellular cytosolic sensors of nucleic acids, including RIG-I and MDA-5, whose role is to detect the presence of pathogenic RNA or DNA within the cell. Ligation of these sensors results in activation of a signaling cassette that ultimately leads to the transcription and secretion of cytokines known as type I interferons. In addition to the nucleic sensors themselves, there are a number of molecules such as TREX1 or RNASEH2B that act as nucleases to degrade pathogenic RNA and DNA and destroy pathogens by eliminating their nucleic acids. These molecules are also important in removing endogenous retroviruses that have been integrated and coadapted into the human genome. Loss of function of the nuclease proteins, or gain of function of the sensors, leads to overactivation of this pathway and the excessive production of type I interferons. It has also been recognized that mutations in the proteasome, a molecular complex responsible for misfolded protein degradation, also results in production of type I interferons.
The major type I interferons include the 13 subclasses of interferon alpha and the two subclasses of interferon beta. All of these molecules signal through the type I interferon receptor (IFNAR). Signaling through IFNAR results in a stereotypical transcriptional program including dozens of genes referred to as interferon regulated genes (IRGs). The collective increase in IRGs after interferon stimulation is often called the interferon signature , and its presence is often taken as a sign of interferon activity even when levels of interferons themselves are unmeasurable. The interferon signature has been found to be associated with a number of autoimmune diseases including systemic lupus erythematosus and dermatomyositis. More recently, a class of inflammatory diseases known as the interferonopathies has been described that result from mutations in the nuclease, RNA sensors, or proteasome molecules described earlier. Because the interferons are a treatable target, by blocking either the cytokine or receptor with a monoclonal antibody, or by blocking IFNAR signaling via a Janus kinase (JAK) inhibitor, mechanistic understanding of this pathway is important for therapeutic development and deployment.
The role of some cytosolic PRRs is complementary to that of membrane-bound TLRs for the activation of inflammatory responses: an example is the role of some NLRP proteins in the activation and secretion of the active form of IL-1β ( Fig. 4.4 ). Unlike most cytokines, IL-1β (together with IL-18) lacks a secretory signal peptide and is externalized through a nonclassical pathway, arranged in two steps. , TLR ligands such as LPS induce gene expression and synthesis of the inactive IL-1β precursor (pro-IL-1β). The protein complex responsible for this catalytic activity is termed the inflammasome. , The inflammasome is composed of the adaptor ASC (apoptosis-associated speck-like protein containing a CARD), procaspase-1, and an NLR family member (such as NLRP1 , NLRP3 , or Ice protease-activating factor [Ipaf]). Oligomerization of these proteins through CARD/CARD interactions results in activation of caspase-1, which cleaves the accumulated IL-1 precursor, resulting in production of biologically active IL-1. ,
A growing number of NLR proteins have been shown to have the capacity to activate caspase 1, each recognizing different danger signals or PAMPs through their respective adaptors , ( Fig. 4.5 ). NLRP3 has been ascribed a role in recognition of adenosine triphosphate (ATP), uric acid crystals, viral RNA, and bacterial DNA. These stimuli are a crucial second hit for the secretion of IL-1β and IL-18 ( Fig. 4.6 ). Indeed, monocytes stimulated with LPS alone release only about 20% of maximal IL-1β. A second stimulus, such as exogenous ATP, strongly enhances proteolytic maturation and secretion of IL-1β. ATP-triggered IL-1β secretion is mediated by P2X7 receptors expressed on the surface of monocytes. Notably, knockout mice deficient in cryopyrin cannot activate caspase-1 upon LPS and ATP stimulation, resulting in lack of IL-1β secretion. In contrast, mutations in the NLRP3 and NLRC4 genes in humans are associated with diseases characterized by excessive production of IL-1β and IL-18, , belonging to the group of the so-called autoinflammatory diseases (see also Chapter 39 ).
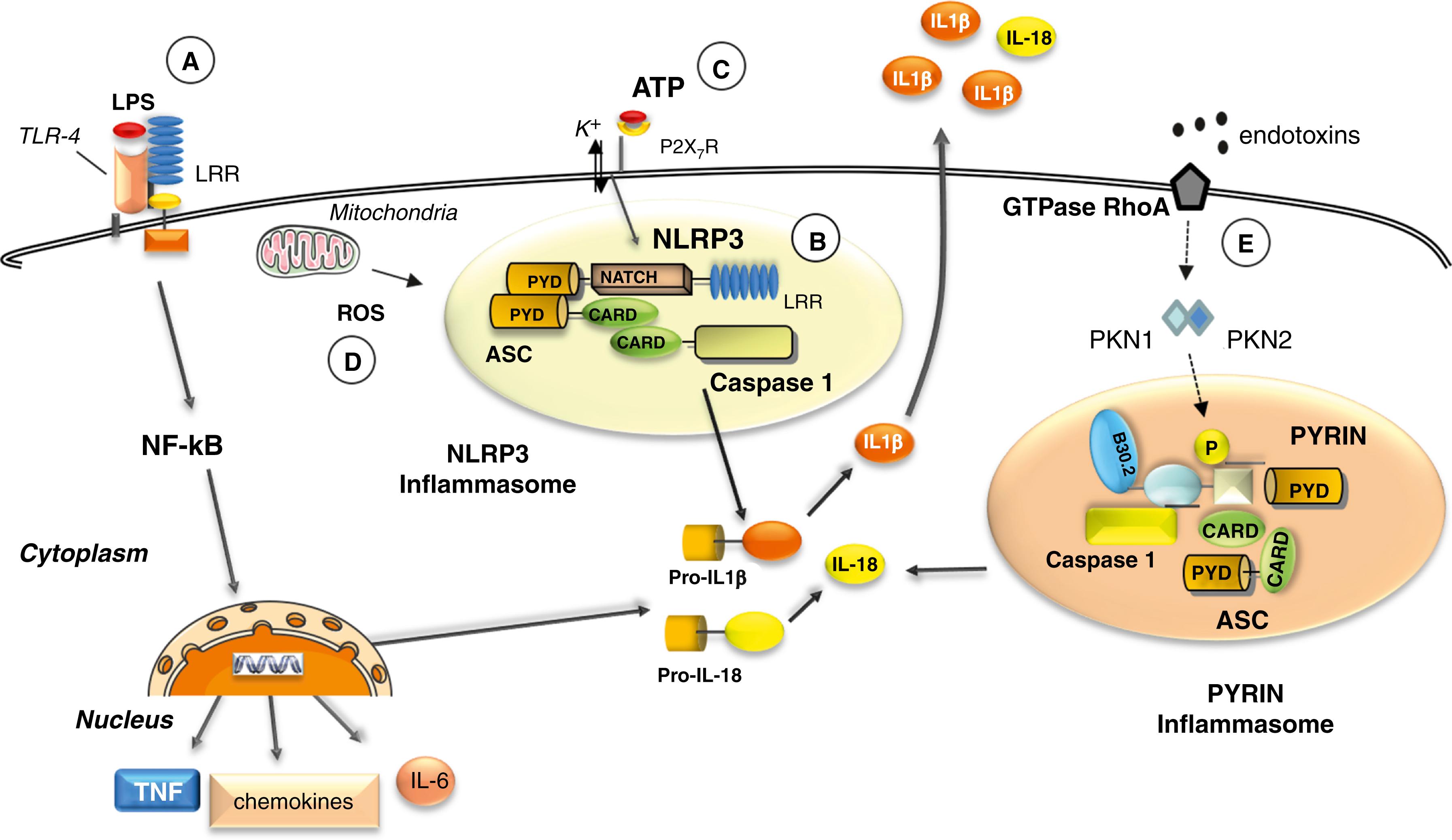
Much evidence suggests a role for oxidative stress in the activation of the NLRP3 -inflammasome. , The exposure of human monocyte to PAMPs and DAMPs induces oxidative stress in the cells through the production of ROS. The extent to which ROS accumulate in the cells is determined by the antioxidant systems that enable cells to maintain redox homeostasis. Under normal conditions, these systems balance the constitutive generation of ROS. Both events —oxidant and antioxidant—are required for the secretion of IL-1β and IL-18 after DAMP or PAMP triggering , ( Fig. 4.6 ). Increased intracellular Ca2+ and decreased cellular cyclic AMP (cAMP) are also able to induce NLRP3 activation through a calcium-sensing receptor (CASR). Ca2+ or other CASR agonists (gadolinium or R-568) activate the NLRP3 inflammasome in the absence of exogenous ATP, whereas knockdown of CASR reduces inflammasome activation in response to known NLRP3 activators. More recently, another route for IL-1β release was demonstrated in mouse macrophages and myelomonocytic cell lines, involving gasdermin-D (GSDMD) and pyroptosis. Infection with intracellular pathogens, mimicked by LPS transfection, activates the noncanonical inflammasome, namely, murine caspase 11 or its human homologs caspase 4/5. In turn, these caspases cleave GSDMD, generating toxin-like peptides that form pores on the plasma membrane mediating the selective secretion of mature IL-1β. This mechanism has been defined as noncanonical inflammasome activation.
Pyrin (or MEFV), the protein mutated in familial Mediterranean fever (FMF), is able to form with ASC and caspase 1 its own “pyrin inflammasome.” In normal conditions, the GTPase RhoA activates the serine-threonine kinases, PKN1 and PKN2, that phosphorylate pyrin. Phosphorylated pyrin binds to regulatory proteins that in turn block the pyrin inflammasome. After exposure to bacterial endotoxins, RhoA GTPase loses its capability to activate PKN1 and PKN2 with the consequent de-phosphorylation of pyrin. Pyrin is therefore able to interact with ASC and caspase 1 inducing IL-1β and IL-18 secretion ( Fig. 4.6 ). Gain of function mutations of MEFV interfere with phosphorylation of pyrin, which is then more prone to activate the pyrin inflammasome upon different stimuli.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here